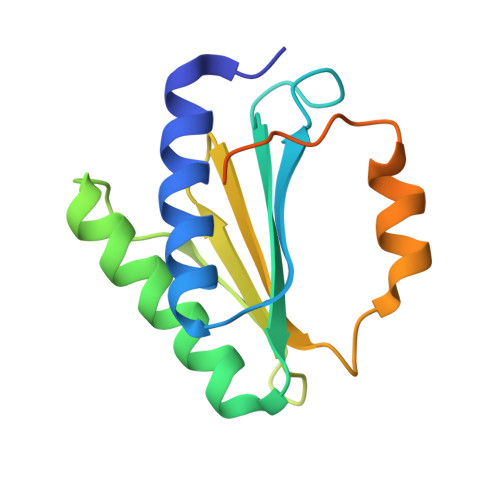
Structure of Ribosomal Silencing Factor Bound to Mycobacterium tuberculosis Ribosome.
Li, X., Sun, Q., Jiang, C., Yang, K., Hung, L.W., Zhang, J., Sacchettini, J.C.(2015) Structure 23: 1858-1865
- PubMed: 26299947
- DOI: https://doi.org/10.1016/j.str.2015.07.014
- Primary Citation of Related Structures:
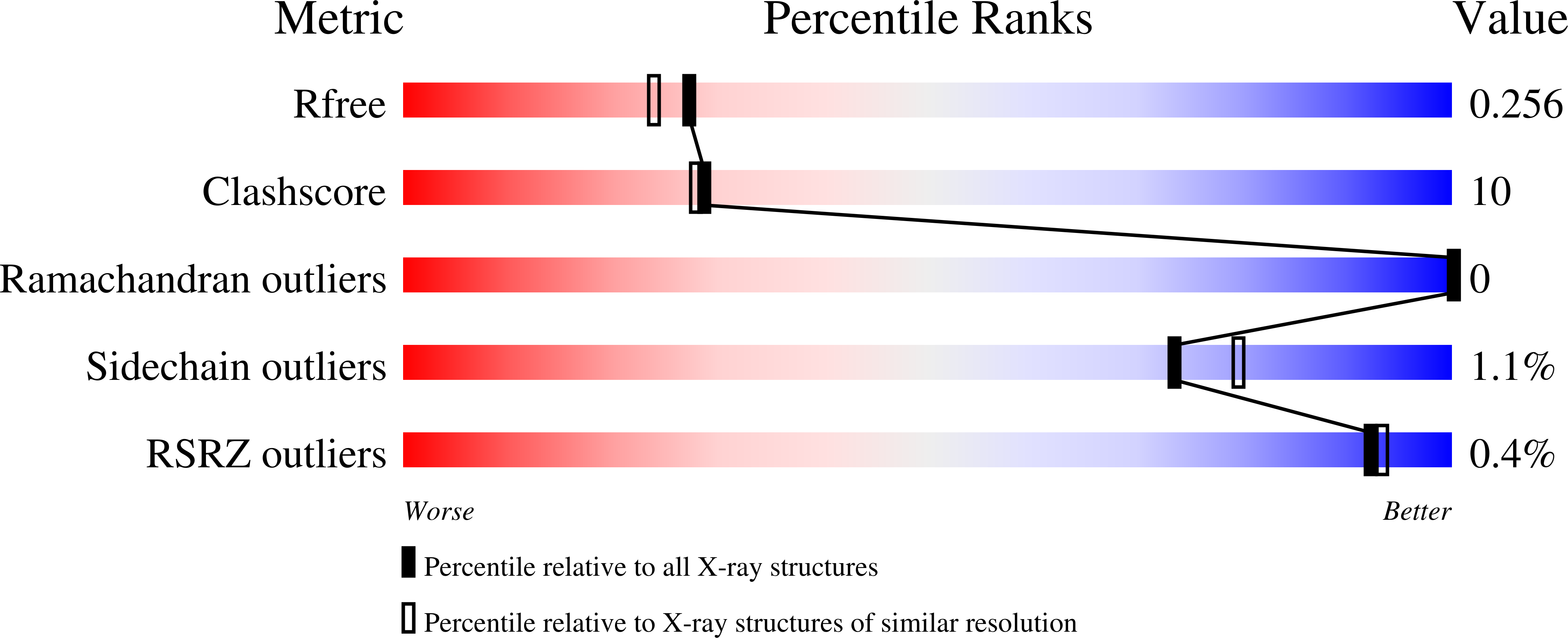
4WCW - PubMed Abstract:
The ribosomal silencing factor RsfS slows cell growth by inhibiting protein synthesis during periods of diminished nutrient availability. The crystal structure of Mycobacterium tuberculosis (Mtb) RsfS, together with the cryo-electron microscopy (EM) structure of the large subunit 50S of Mtb ribosome, reveals how inhibition of protein synthesis by RsfS occurs. RsfS binds to the 50S at L14, which, when occupied, blocks the association of the small subunit 30S. Although Mtb RsfS is a dimer in solution, only a single subunit binds to 50S. The overlap between the dimer interface and the L14 binding interface confirms that the RsfS dimer must first dissociate to a monomer in order to bind to L14. RsfS interacts primarily through electrostatic and hydrogen bonding to L14. The EM structure shows extended rRNA density that it is not found in the Escherichia coli ribosome, the most striking of these being the extended RNA helix of H54a.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843, USA.