The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling.
Hsu, F., Luo, X., Qiu, J., Teng, Y.B., Jin, J., Smolka, M.B., Luo, Z.Q., Mao, Y.(2014) Proc Natl Acad Sci U S A 111: 10538-10543
- PubMed: 25006264
- DOI: https://doi.org/10.1073/pnas.1402605111
- Primary Citation of Related Structures:
4TRG, 4TRH - PubMed Abstract:
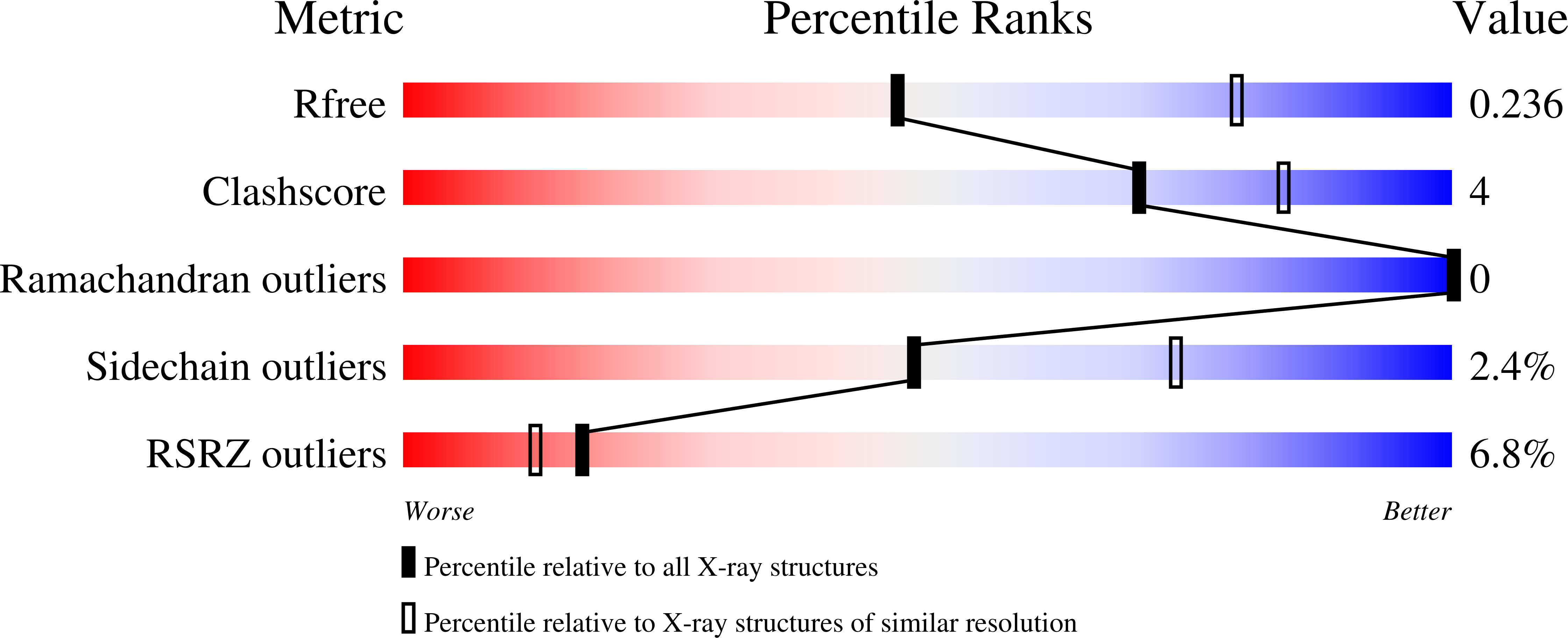
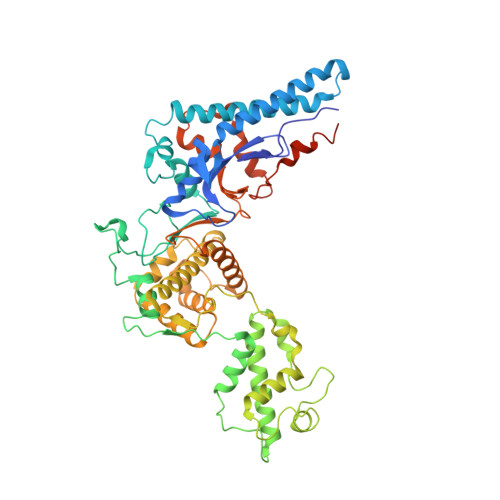
The activity of proteins delivered into host cells by the Dot/Icm injection apparatus allows Legionella pneumophila to establish a niche called the Legionella-containing vacuole (LCV), which is permissive for intracellular bacterial propagation. Among these proteins, substrate of Icm/Dot transporter (SidC) anchors to the cytoplasmic surface of the LCV and is important for the recruitment of host endoplasmic reticulum (ER) proteins to this organelle. However, the biochemical function underlying this activity is unknown. Here, we determined the structure of the N-terminal domain of SidC, which has no structural homology to any protein. Sequence homology analysis revealed a potential canonical catalytic triad formed by Cys46, His444, and Asp446 on the surface of SidC. Unexpectedly, we found that SidC is an E3 ubiquitin ligase that uses the C-H-D triad to catalyze the formation of high-molecular-weight polyubiquitin chains through multiple ubiquitin lysine residues. A C46A mutation completely abolished the E3 ligase activity and the ability of the protein to recruit host ER proteins as well as polyubiquitin conjugates to the LCV. Thus, SidC represents a unique E3 ubiquitin ligase family important for phagosomal membrane remodeling by L. pneumophila.
Organizational Affiliation:
Weill Institute for Cell and Molecular Biology and Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853;