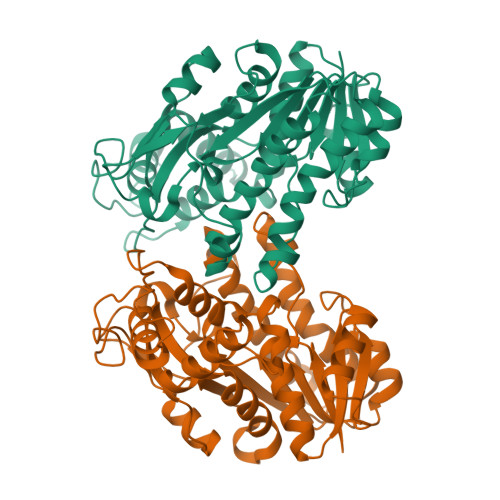
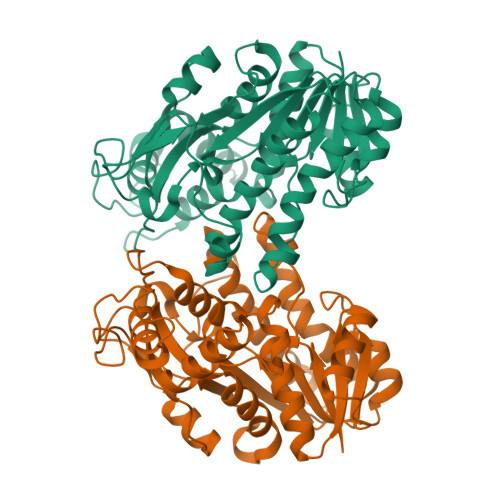
Structural insights into the cofactor-assisted substrate recognition of yeast methylglyoxal/isovaleraldehyde reductase Gre2
Guo, P.C., Bao, Z.Z., Ma, X.X., Xia, Q., Li, W.F.(2014) Biochim Biophys Acta 1844: 1486-1492
- PubMed: 24879127
- DOI: https://doi.org/10.1016/j.bbapap.2014.05.008
- Primary Citation of Related Structures:
4PVC, 4PVD - PubMed Abstract:
Saccharomyces cerevisiae Gre2 (EC1.1.1.283) serves as a versatile enzyme that catalyzes the stereoselective reduction of a broad range of substrates including aliphatic and aromatic ketones, diketones, as well as aldehydes, using NADPH as the cofactor. Here we present the crystal structures of Gre2 from S. cerevisiae in an apo-form at 2.00Å and NADPH-complexed form at 2.40Å resolution. Gre2 forms a homodimer, each subunit of which contains an N-terminal Rossmann-fold domain and a variable C-terminal domain, which participates in substrate recognition. The induced fit upon binding to the cofactor NADPH makes the two domains shift toward each other, producing an interdomain cleft that better fits the substrate. Computational simulation combined with site-directed mutagenesis and enzymatic activity analysis enabled us to define a potential substrate-binding pocket that determines the stringent substrate stereoselectivity for catalysis.
Organizational Affiliation:
State Key Laboratory of Silkworm Genome Biology, Southwest University, 216, Tiansheng Road, Beibei, Chongqing 400716, People's Republic of China; Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, People's Republic of China.