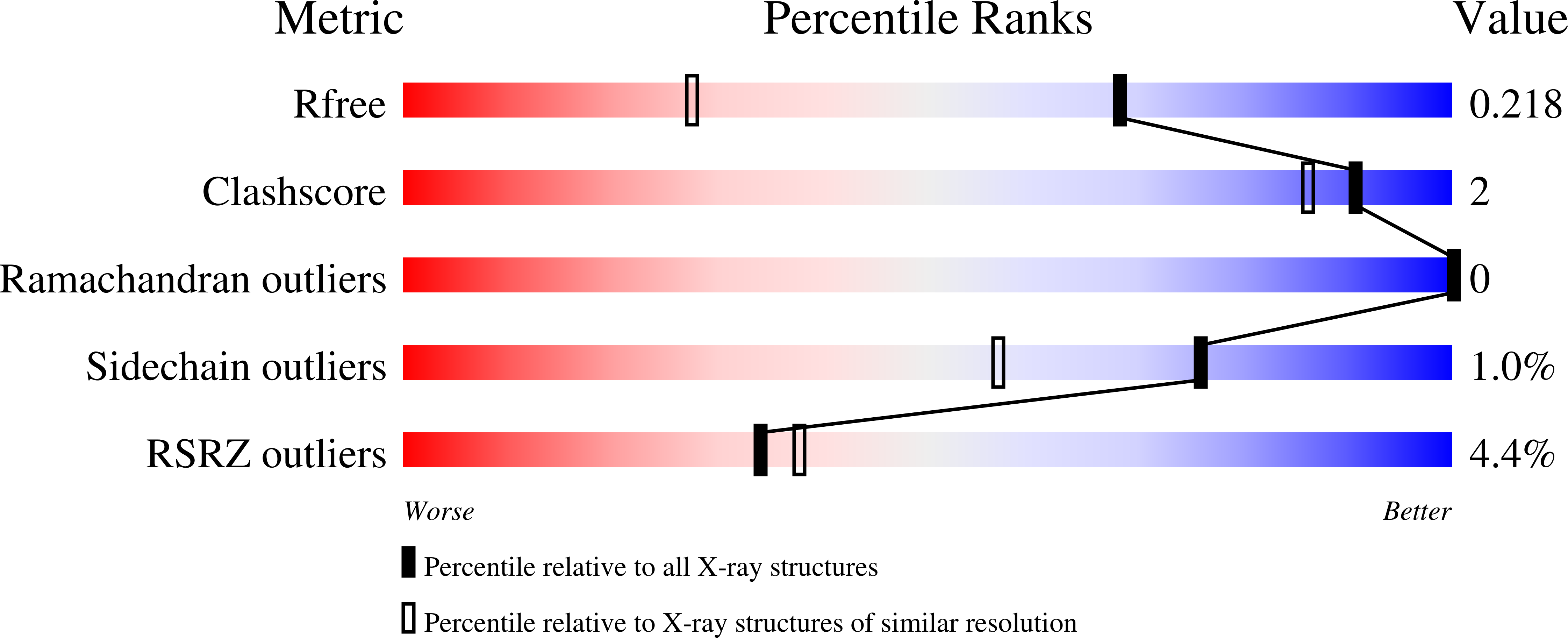
The Pseudomonas aeruginosa phosphate transport protein PstS plays a phosphate-independent role in biofilm formation.
Neznansky, A., Blus-Kadosh, I., Yerushalmi, G., Banin, E., Opatowsky, Y.(2014) FASEB J 28: 5223-5233
- PubMed: 25223609
- DOI: https://doi.org/10.1096/fj.14-258293
- Primary Citation of Related Structures:
4OMB - PubMed Abstract:
Pseudomonas aeruginosa (PA) is a primary cause of nosocomial infections. A key element in PA pathogenicity is its ability to form biofilms that withstand eradication by antibiotics and the immune system. Biofilm formation is controlled by phosphate signaling and here we provide evidence that PstS, a subunit of the PA Pst phosphate transporter, has a surprising role in this process. Using X-ray crystallography, we characterized the unique underpinnings of PstS phosphate binding and identified an unusual 15-residue N' loop extension. Structure-based experiments showed that PstS-mediated phosphate uptake and biofilm formation are in fact two distinct functions. Specifically, a point mutation that abrogated phosphate binding did not eliminate biofilm formation; conversely, truncation of the N' loop diminished the ability of PA to form biofilms but had no effect on phosphate binding and uptake. This places PstS at a junction that separately controls phosphate sensing and uptake and the ultrastructure organization of bacteria.
Organizational Affiliation:
Mina and Everard Goodman Faculty of Life Sciences and.