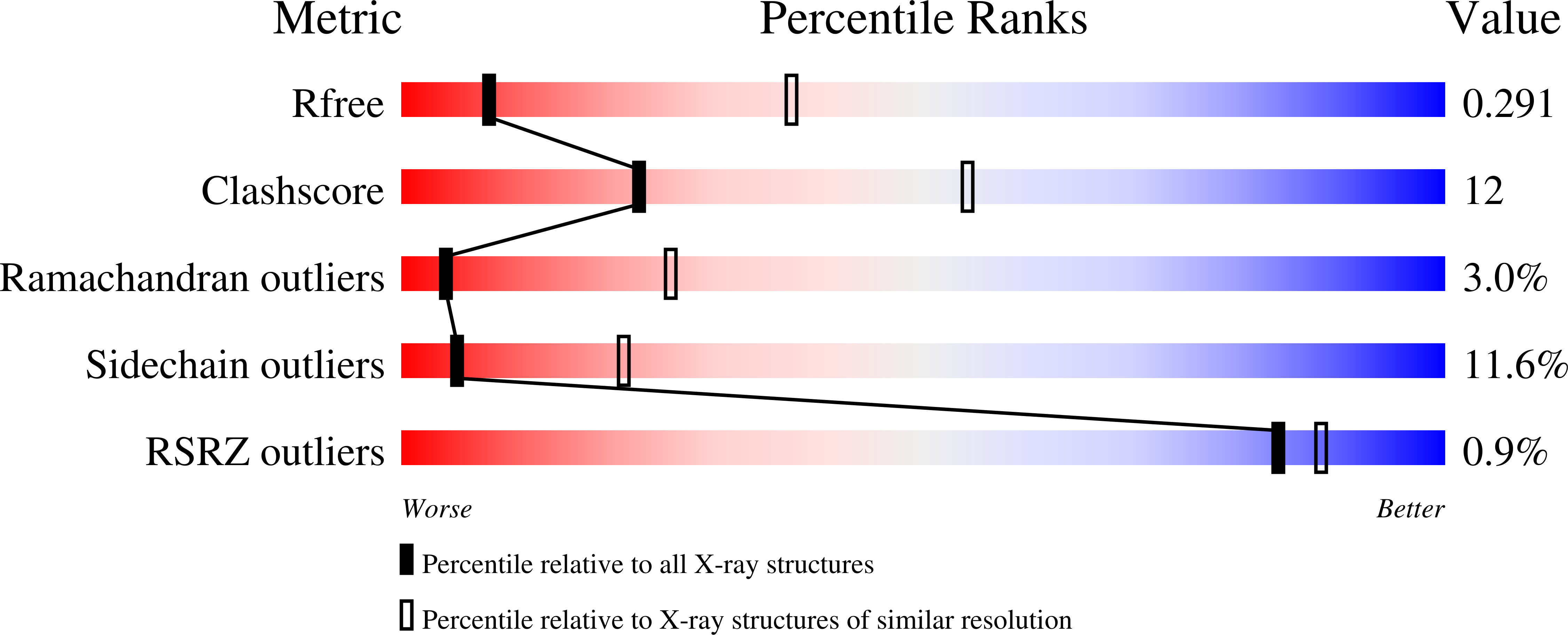
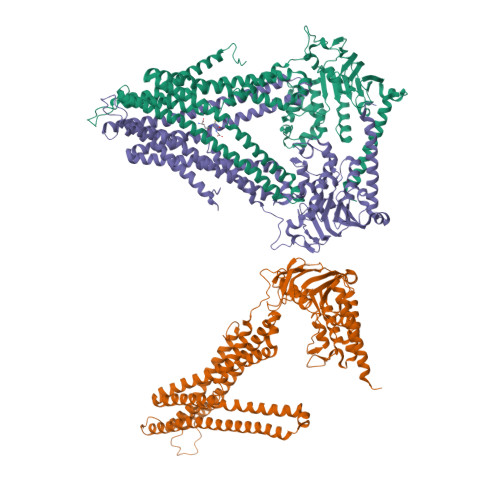
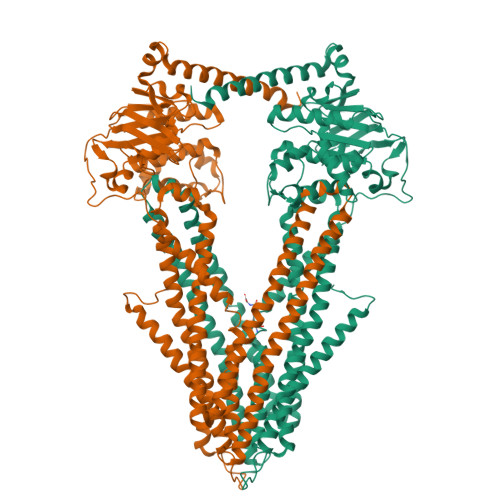
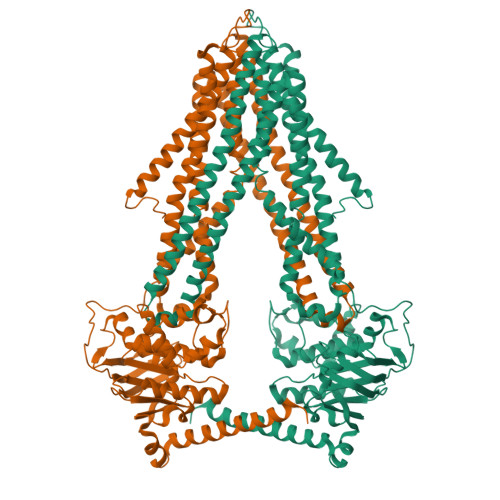
Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1.
Srinivasan, V., Pierik, A.J., Lill, R.(2014) Science 343: 1137-1140
- PubMed: 24604199
- DOI: https://doi.org/10.1126/science.1246729
- Primary Citation of Related Structures:
4MYC, 4MYH - PubMed Abstract:
The yeast mitochondrial ABC transporter Atm1, in concert with glutathione, functions in the export of a substrate required for cytosolic-nuclear iron-sulfur protein biogenesis and cellular iron regulation. Defects in the human ortholog ABCB7 cause the sideroblastic anemia XLSA/A. Here, we report the crystal structures of free and glutathione-bound Atm1 in inward-facing, open conformations at 3.06- and 3.38-angstrom resolution, respectively. The glutathione binding site includes a residue mutated in XLSA/A and is located close to the inner membrane surface in a large cavity. The two nucleotide-free adenosine 5'-triphosphate binding domains do not interact yet are kept in close vicinity through tight interaction of the two C-terminal α-helices of the Atm1 dimer. The resulting protein stabilization may be a common structural feature of all ABC exporters.
Organizational Affiliation:
Institut für Zytobiologie, Philipps-Universität Marburg, Robert-Koch-Strasse 6, 35032 Marburg, Germany.