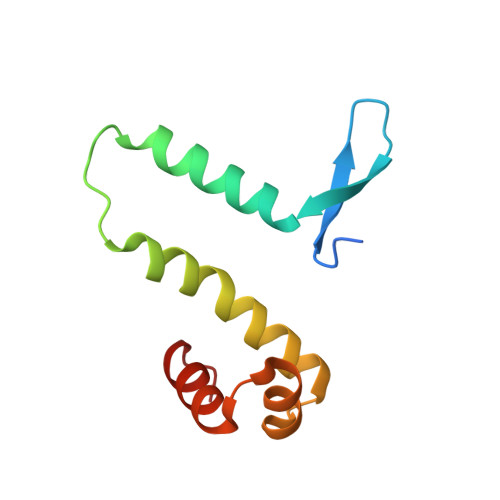
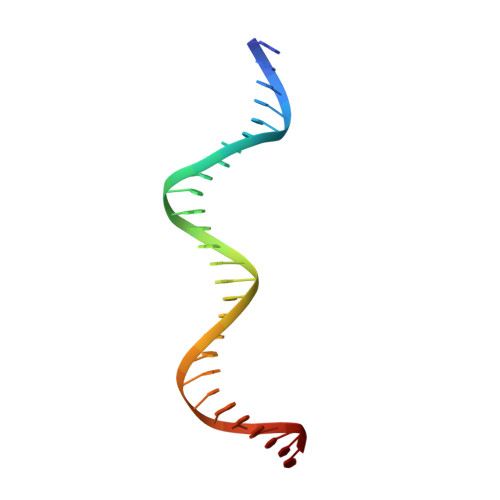
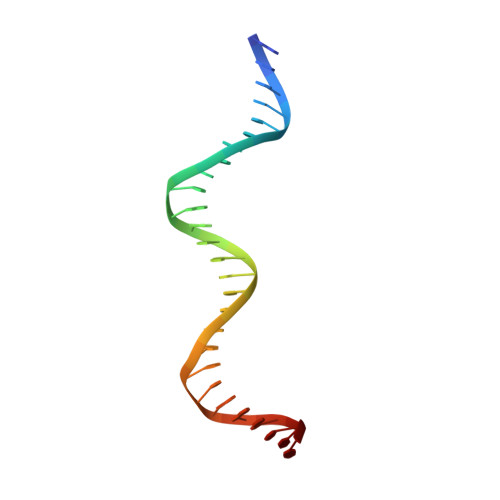
Control of DNA minor groove width and Fis protein binding by the purine 2-amino group.
Hancock, S.P., Ghane, T., Cascio, D., Rohs, R., Di Felice, R., Johnson, R.C.(2013) Nucleic Acids Res 41: 6750-6760
- PubMed: 23661683
- DOI: https://doi.org/10.1093/nar/gkt357
- Primary Citation of Related Structures:
4IHV, 4IHW, 4IHX, 4IHY - PubMed Abstract:
The width of the DNA minor groove varies with sequence and can be a major determinant of DNA shape recognition by proteins. For example, the minor groove within the center of the Fis-DNA complex narrows to about half the mean minor groove width of canonical B-form DNA to fit onto the protein surface. G/C base pairs within this segment, which is not contacted by the Fis protein, reduce binding affinities up to 2000-fold over A/T-rich sequences. We show here through multiple X-ray structures and binding properties of Fis-DNA complexes containing base analogs that the 2-amino group on guanine is the primary molecular determinant controlling minor groove widths. Molecular dynamics simulations of free-DNA targets with canonical and modified bases further demonstrate that sequence-dependent narrowing of minor groove widths is modulated almost entirely by the presence of purine 2-amino groups. We also provide evidence that protein-mediated phosphate neutralization facilitates minor groove compression and is particularly important for binding to non-optimally shaped DNA duplexes.
Organizational Affiliation:
Department of Biological Chemistry, David Geffen School of Medicine at the University of California at Los Angeles, Los Angeles, CA 90095-1737, USA.