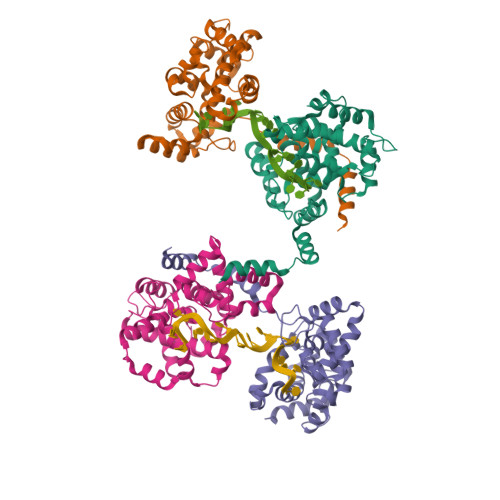
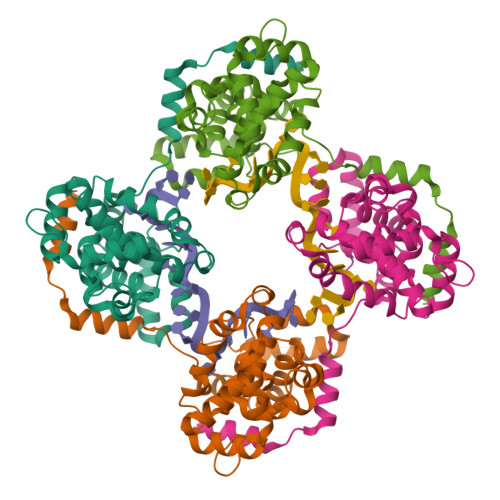
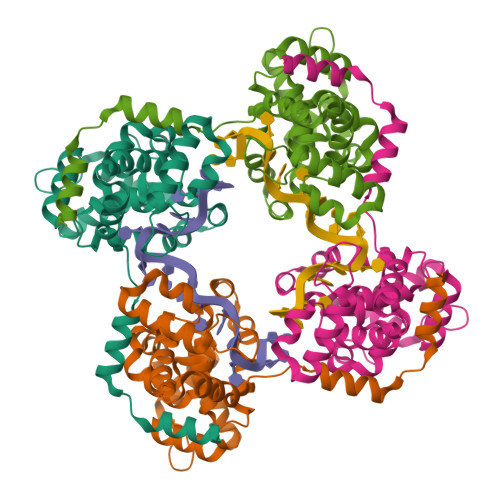
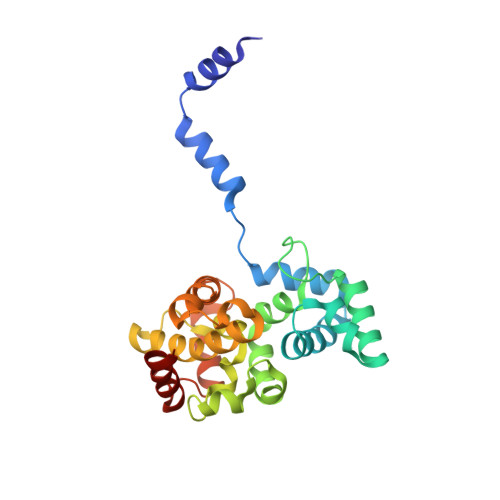
Phleboviruses encapsidate their genomes by sequestering RNA bases.
Raymond, D.D., Piper, M.E., Gerrard, S.R., Skiniotis, G., Smith, J.L.(2012) Proc Natl Acad Sci U S A 109: 19208-19213
- PubMed: 23129612
- DOI: https://doi.org/10.1073/pnas.1213553109
- Primary Citation of Related Structures:
4H5L, 4H5M, 4H5O, 4H5P, 4H5Q, 4V9E - PubMed Abstract:
Rift Valley fever and Toscana viruses are human pathogens for which no effective therapeutics exist. These and other phleboviruses have segmented negative-sense RNA genomes that are sequestered by a nucleocapsid protein (N) to form ribonucleoprotein (RNP) complexes of irregular, asymmetric structure, previously uncharacterized at high resolution. N binds nonspecifically to single-stranded RNA with nanomolar affinity. Crystal structures of Rift Valley fever virus N-RNA complexes reconstituted with defined RNAs of different length capture tetrameric, pentameric and hexameric N-RNA multimers. All N-N subunit contacts are mediated by a highly flexible α-helical arm. Arm movement gives rise to the three multimers in the crystal structures and also explains the asymmetric architecture of the RNP. Despite the flexible association of subunits, the crystal structures reveal an invariant, monomeric RNP building block, consisting of the core of one N subunit, the arm of a neighboring N, and four RNA nucleotides with the flanking phosphates. Up to three additional RNA nucleotides bind between subunits. The monomeric building block is matched in size to the repeating unit in viral RNP, as visualized by electron microscopy. N sequesters four RNA bases in a narrow hydrophobic binding slot and has polar contacts only with the sugar-phosphate backbone, which faces the solvent. All RNA bases, whether in the binding slot or in the subunit interface, face the protein in a manner that is incompatible with base pairing or with "reading" by the viral polymerase.
Organizational Affiliation:
Life Sciences Institute, Department of Biological Chemistry, Cell and Molecular Biology Program, University of Michigan, Ann Arbor, MI 48109, USA.