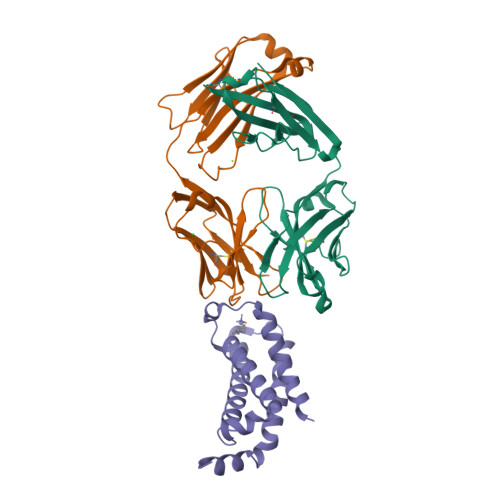
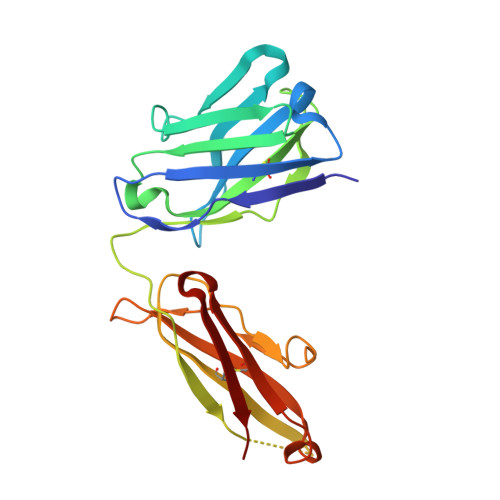
Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain.
Li, Q., Wanderling, S., Paduch, M., Medovoy, D., Singharoy, A., McGreevy, R., Villalba-Galea, C.A., Hulse, R.E., Roux, B., Schulten, K., Kossiakoff, A., Perozo, E.(2014) Nat Struct Mol Biol 21: 244-252
- PubMed: 24487958
- DOI: https://doi.org/10.1038/nsmb.2768
- Primary Citation of Related Structures:
4G7V, 4G7Y, 4G80 - PubMed Abstract:
The transduction of transmembrane electric fields into protein motion has an essential role in the generation and propagation of cellular signals. Voltage-sensing domains (VSDs) carry out these functions through reorientations of positive charges in the S4 helix. Here, we determined crystal structures of the Ciona intestinalis VSD (Ci-VSD) in putatively active and resting conformations. S4 undergoes an ~5-Å displacement along its main axis, accompanied by an ~60° rotation. This movement is stabilized by an exchange in countercharge partners in helices S1 and S3 that generates an estimated net charge transfer of ~1 eo. Gating charges move relative to a ''hydrophobic gasket' that electrically divides intra- and extracellular compartments. EPR spectroscopy confirms the limited nature of S4 movement in a membrane environment. These results provide an explicit mechanism for voltage sensing and set the basis for electromechanical coupling in voltage-dependent enzymes and ion channels.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, Illinois, USA.