Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Deng, L., Langley, R.J., Wang, Q., Topalian, S.L., Mariuzza, R.A.(2012) Proc Natl Acad Sci U S A
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2012) Proc Natl Acad Sci U S A
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HLA class II histocompatibility antigen, DR alpha chain | 182 | Homo sapiens | Mutation(s): 0 Gene Names: HLA-DRA, HLA-DRA1 | 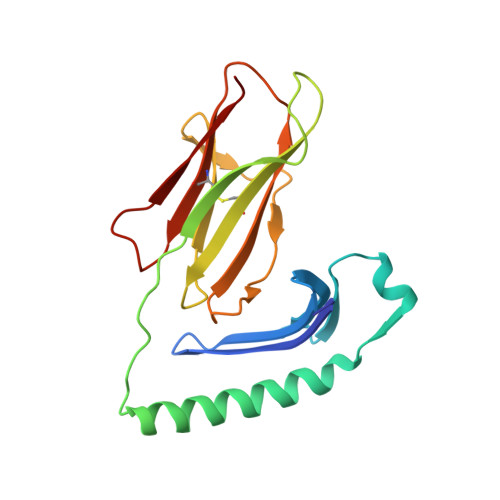 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01903 (Homo sapiens) Explore P01903 Go to UniProtKB: P01903 | |||||
PHAROS: P01903 GTEx: ENSG00000204287 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01903 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HLA class II histocompatibility antigen, DRB1-1 beta chain | 190 | Homo sapiens | Mutation(s): 0 Gene Names: HLA-DRB1 | 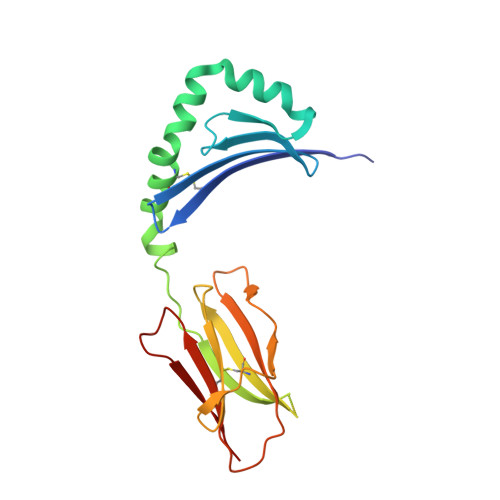 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01911 (Homo sapiens) Explore P01911 Go to UniProtKB: P01911 | |||||
PHAROS: P01911 GTEx: ENSG00000196126 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01911 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Triosephosphate isomerase | 15 | Homo sapiens | Mutation(s): 0 Gene Names: TPI1, TPI EC: 5.3.1.1 (PDB Primary Data), 4.2.3.3 (UniProt) | 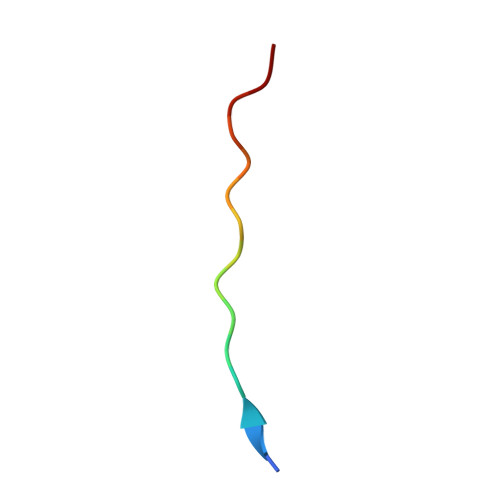 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P60174 (Homo sapiens) Explore P60174 Go to UniProtKB: P60174 | |||||
PHAROS: P60174 GTEx: ENSG00000111669 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60174 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T cell receptor G4 alpha chain | 203 | Homo sapiens | Mutation(s): 0 | 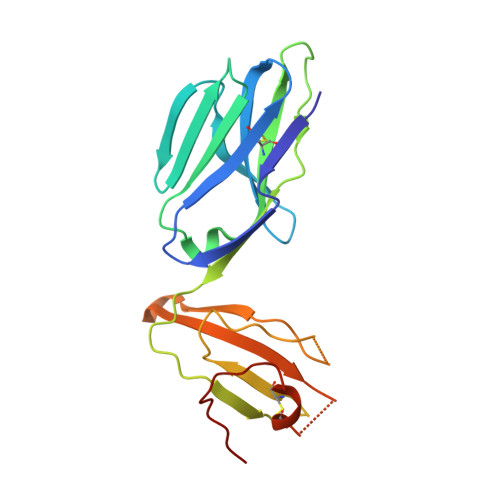 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T cell receptor G4 beta chain | 239 | Homo sapiens | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NA Query on NA | K [auth A], L [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 126.133 | α = 90 |
| b = 175.611 | β = 110.75 |
| c = 88.646 | γ = 90 |
| Software Name | Purpose |
|---|---|
| ADSC | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |