The Epithelial Adhesin 1 (Epa1P) from the Human-Pathogenic Yeast Candida Glabrata : Structural and Functional Study of the Carbohydrate-Binding Domain
Ielasi, F.S., Decanniere, K., Willaert, R.G.(2012) Acta Crystallogr D Biol Crystallogr 68: 210
- PubMed: 22349222
- DOI: https://doi.org/10.1107/S0907444911054898
- Primary Citation of Related Structures:
4A3X - PubMed Abstract:
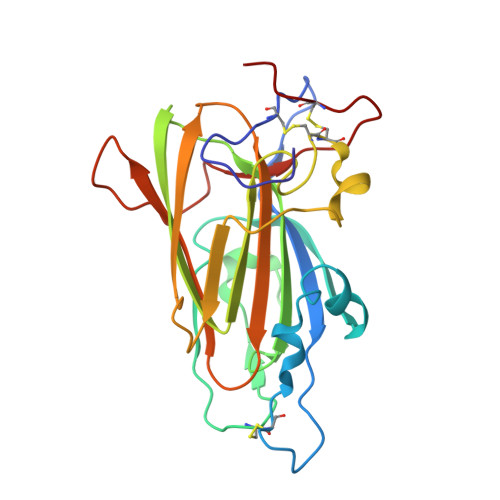
The yeast Candida glabrata represents the second major cause of clinical candidiasis cases in the world. The ability of this opportunistic pathogen to adhere to human epithelial and endothelial cells relies on the Epa adhesins, a large set of cell-wall proteins whose N-terminal domains are endowed with a calcium-dependent lectin activity. This feature allows the yeast cells to adhere to host cells by establishing multiple interactions with the glycans expressed on their cell membrane. The ligand-binding domain of the Epa1p adhesin, which is one of the best characterized in the Epa family, was expressed in Escherichia coli, purified and crystallized in complex with lactose. Sequence identity with the domain of another yeast adhesin, the Flo5p flocculin from Saccharomyces cerevisiae, was exploited for molecular replacement and the structure of the domain was solved at a resolution of 1.65 Å. The protein is a member of the PA14 superfamily. It has a β-sandwich core and a DcisD calcium-binding motif, which is also present in the binding site of Flo5p. However, Epa1p differs from this homologue by the lack of a Flo5-like subdomain and by a significantly decreased accessibility of the solvent to the binding site, in which a calcium ion still plays an active role in the interactions with carbohydrates. This structural insight, together with fluorescence-assay data, confirms and explains the higher specificity of Epa1p adhesin for glycan molecules compared with the S. cerevisiae flocculins.
Organizational Affiliation:
Research Group Structural Biology Brussels (SBB), Department of Bioengineering Sciences, Vrije Universiteit Brussel, Pleinlaan 2, 1050 Brussels, Belgium.