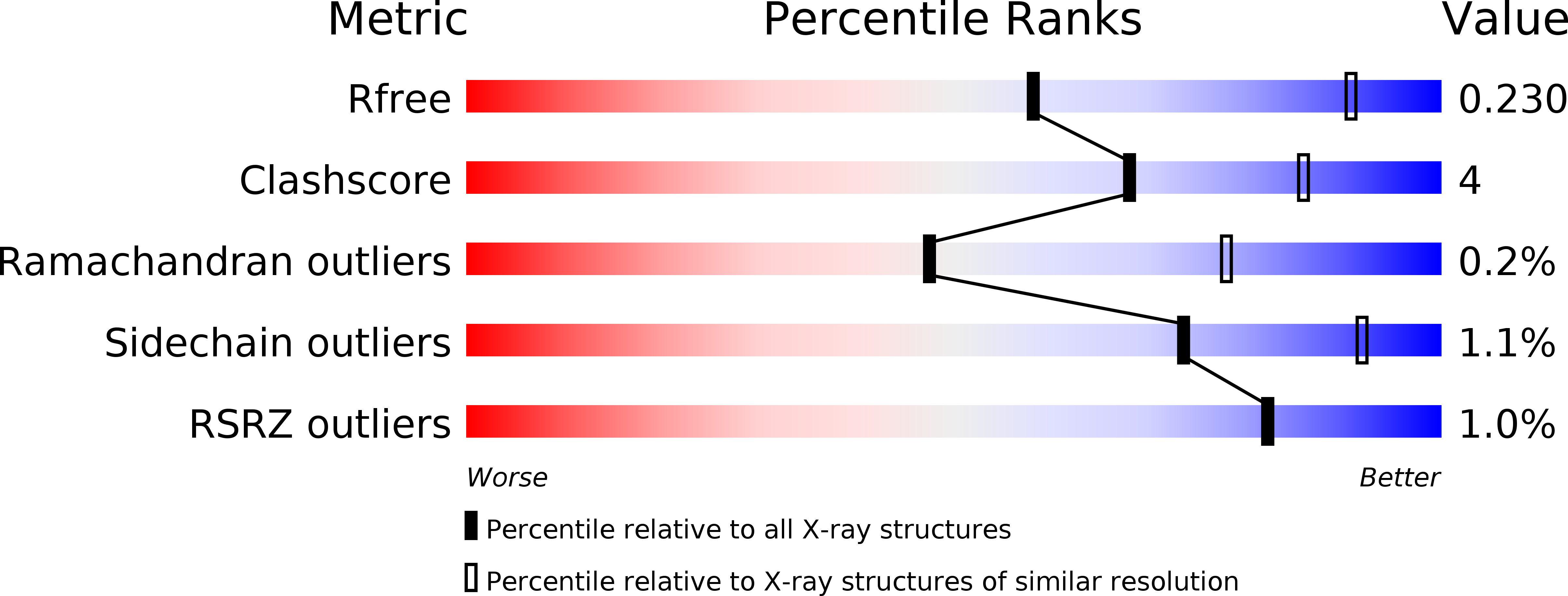
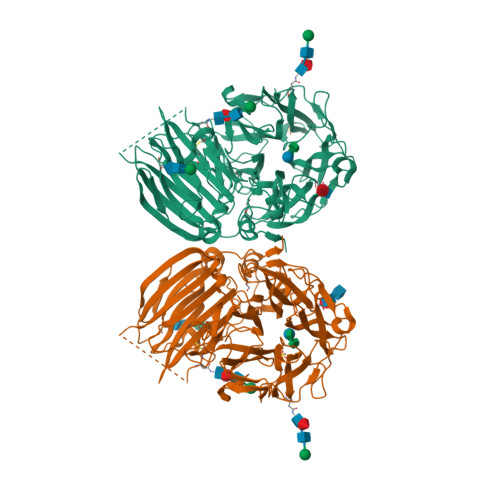
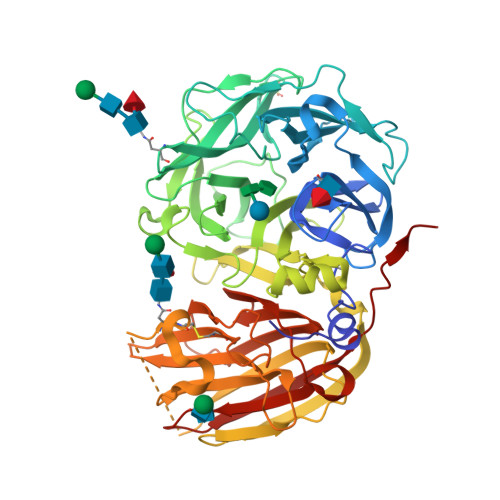
Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Lammens, W., Le Roy, K., Yuan, S., Vergauwen, R., Rabijns, A., Van Laere, A., Strelkov, S.V., Van den Ende, W.(2012) Plant J 70: 205-219
- PubMed: 22098191
- DOI: https://doi.org/10.1111/j.1365-313X.2011.04858.x
- Primary Citation of Related Structures:
3UGF, 3UGG, 3UGH - PubMed Abstract:
Fructans play important roles as reserve carbohydrates and stress protectants in plants, and additionally serve as prebiotics with emerging antioxidant properties. Various fructan types are synthesized by an array of plant fructosyltransferases belonging to family 32 of the glycoside hydrolases (GH32), clustering together with GH68 in Clan-J. Here, the 3D structure of a plant fructosyltransferase from a native source, the Pachysandra terminalis 6-SST/6-SFT (Pt6-SST/6-SFT), is reported. In addition to its 1-SST (1-kestose-forming) and hydrolytic side activities, the enzyme uses sucrose to create graminan- and levan-type fructans, which are probably associated with cold tolerance in this species. Furthermore, a Pt6-SST/6-SFT complex with 6-kestose was generated, representing a genuine acceptor binding modus at the +1, +2 and +3 subsites in the active site. The enzyme shows a unique configuration in the vicinity of its active site, including a unique D/Q couple located at the +1 subsite that plays a dual role in donor and acceptor substrate binding. Furthermore, it shows a unique orientation of some hydrophobic residues, probably contributing to its specific functionality. A model is presented showing formation of a β(2-6) fructosyl linkage on 6-kestose to create 6,6-nystose, a mechanism that differs from the creation of a β(2-1) fructosyl linkage on sucrose to produce 1-kestose. The structures shed light on the evolution of plant fructosyltransferases from their vacuolar invertase ancestors, and contribute to further understanding of the complex structure-function relationships within plant GH32 members.
Organizational Affiliation:
Biology Department, Laboratory for Molecular Plant Physiology, Katholieke Universiteit Leuven, Kasteelpark Arenberg 31, Box 2434, B-3001 Heverlee, Belgium.