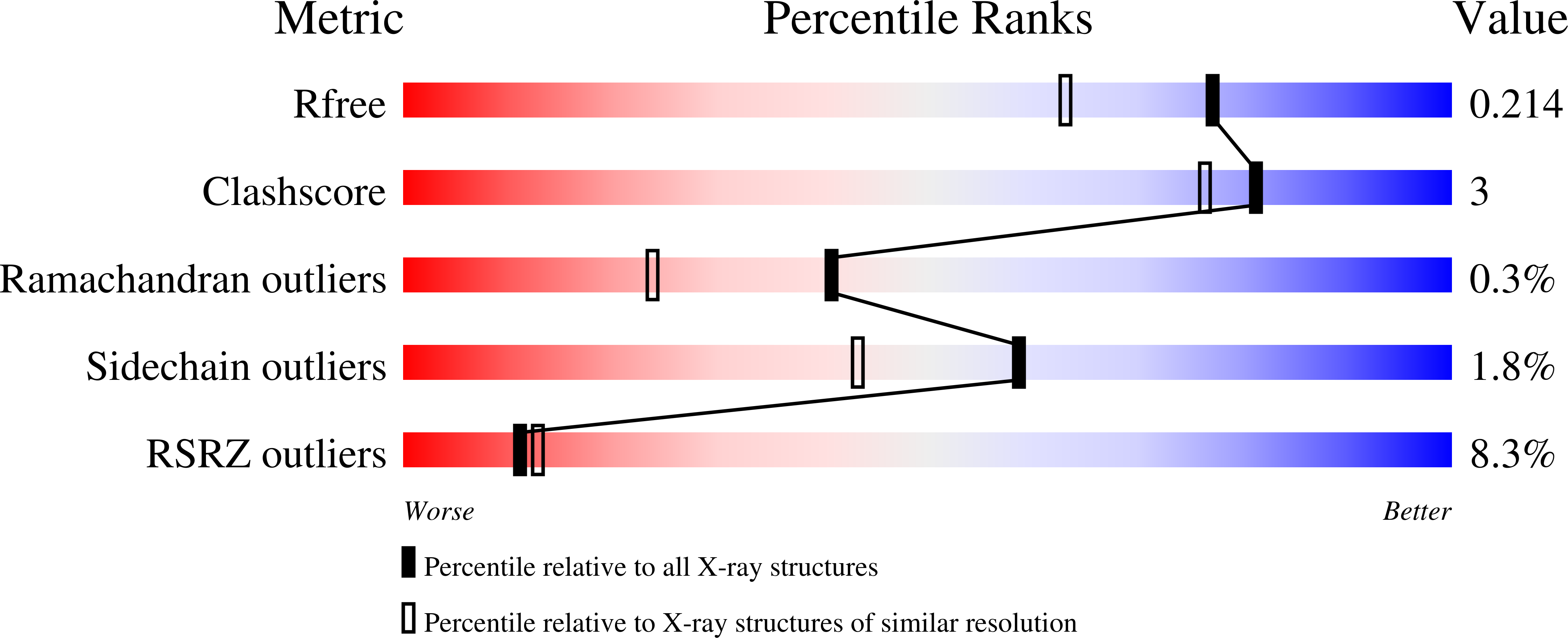
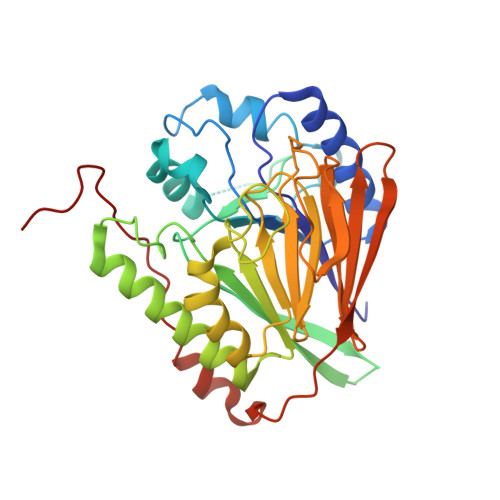
Crystal structure of GAP50, the anchor of the invasion machinery in the inner membrane complex of Plasmodium falciparum.
Bosch, J., Paige, M.H., Vaidya, A.B., Bergman, L.W., Hol, W.G.(2012) J Struct Biol 178: 61-73
- PubMed: 22387043
- DOI: https://doi.org/10.1016/j.jsb.2012.02.009
- Primary Citation of Related Structures:
3TGH - PubMed Abstract:
The glideosome associated protein GAP50 is an essential protein in apicomplexan parasites such as Plasmodium, Toxoplasma and Cryptosporidium, several species of which are important human pathogens. The 44.6kDa protein is part of a multi-protein complex known as the invasion machinery or glideosome, which is required for cell invasion and substrate gliding motility empowered by an actin-myosin motor. GAP50 is anchored through its C-terminal transmembrane helix into the inner membrane complex and interacts via a short six residue C-terminal tail with other proteins of the invasion machinery in the pellicle of the parasite. In this paper we describe the 1.7Å resolution crystal structure of the soluble GAP50 domain from the malaria parasite Plasmodium falciparum. The structure shows an αßßα fold with overall similarity to purple acid phosphatases with, however, little homology regarding the nature of the residues in the active site region of the latter enzyme. While purple acid phosphatases contain a phosphate bridged binuclear Fe-site coordinated by seven side chains with the Fe-ions 3.2Å apart, GAP50 in our crystals contains two cobalt ions each with one protein ligand and a distance between the Co(2+) ions of 18Å.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, WA 98195, USA.