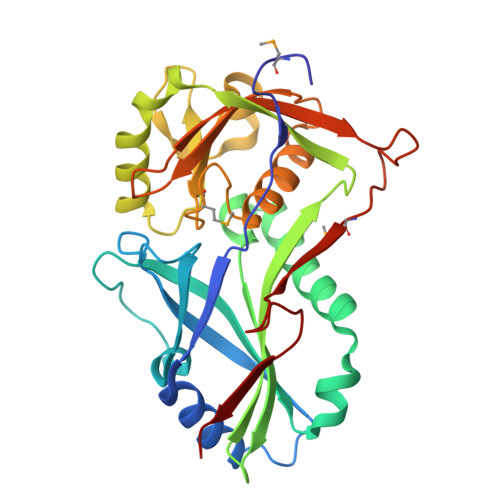
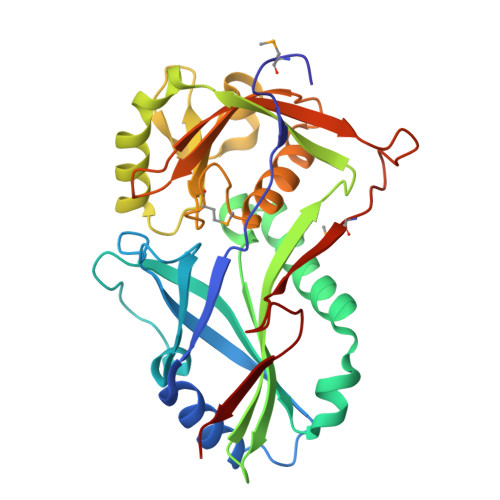
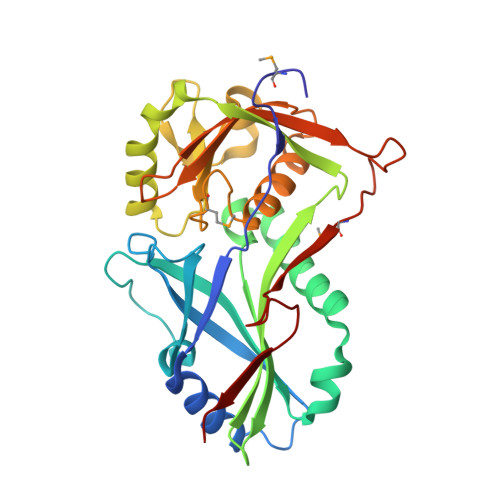
Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Taherbhoy, A.M., Tait, S.W., Kaiser, S.E., Williams, A.H., Deng, A., Nourse, A., Hammel, M., Kurinov, I., Rock, C.O., Green, D.R., Schulman, B.A.(2011) Mol Cell 44: 451-461
- PubMed: 22055190
- DOI: https://doi.org/10.1016/j.molcel.2011.08.034
- Primary Citation of Related Structures:
3T7E, 3T7F, 3T7G, 3T7H - PubMed Abstract:
Atg7 is a noncanonical, homodimeric E1 enzyme that interacts with the noncanonical E2 enzyme, Atg3, to mediate conjugation of the ubiquitin-like protein (UBL) Atg8 during autophagy. Here we report that the unique N-terminal domain of Atg7 (Atg7(NTD)) recruits a unique "flexible region" from Atg3 (Atg3(FR)). The structure of an Atg7(NTD)-Atg3(FR) complex reveals hydrophobic residues from Atg3 engaging a conserved groove in Atg7, important for Atg8 conjugation. We also report the structure of the homodimeric Atg7 C-terminal domain, which is homologous to canonical E1s and bacterial antecedents. The structures, SAXS, and crosslinking data allow modeling of a full-length, dimeric (Atg7~Atg8-Atg3)(2) complex. The model and biochemical data provide a rationale for Atg7 dimerization: Atg8 is transferred in trans from the catalytic cysteine of one Atg7 protomer to Atg3 bound to the N-terminal domain of the opposite Atg7 protomer within the homodimer. The studies reveal a distinctive E1~UBL-E2 architecture for enzymes mediating autophagy.
Organizational Affiliation:
Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN 38105, USA.