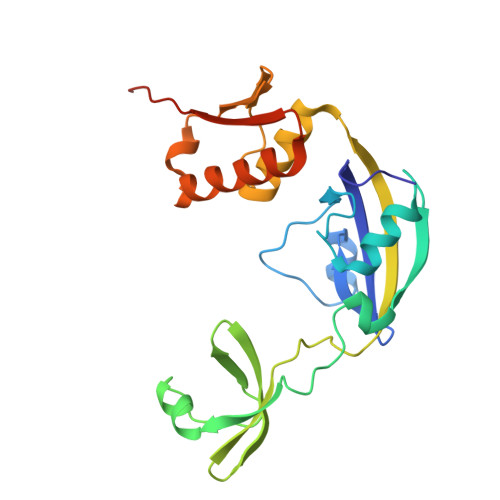
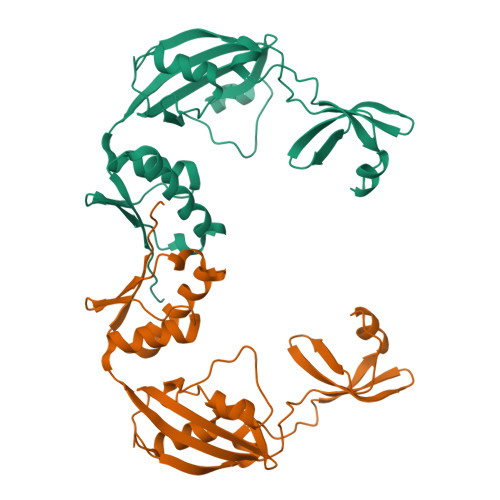
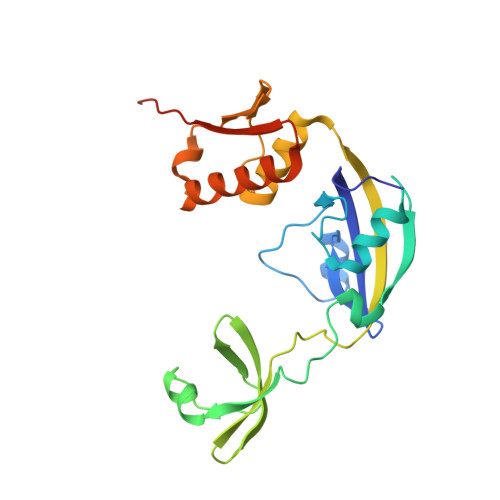
Structural Analysis of Protein Folding by the Long-Chain Archaeal Chaperone FKBP26.
Martinez-Hackert, E., Hendrickson, W.A.(2011) J Mol Biology 407: 450-464
- PubMed: 21262232
- DOI: https://doi.org/10.1016/j.jmb.2011.01.027
- Primary Citation of Related Structures:
3PR9, 3PRA, 3PRB, 3PRD - PubMed Abstract:
In the cell, protein folding is mediated by folding catalysts and chaperones. The two functions are often linked, especially when the catalytic module forms part of a multidomain protein, as in Methanococcus jannaschii peptidyl-prolyl cis/trans isomerase FKBP26. Here, we show that FKBP26 chaperone activity requires both a 50-residue insertion in the catalytic FKBP domain, also called 'Insert-in-Flap' or IF domain, and an 80-residue C-terminal domain. We determined FKBP26 structures from four crystal forms and analyzed chaperone domains in light of their ability to mediate protein-protein interactions. FKBP26 is a crescent-shaped homodimer. We reason that folding proteins are bound inside the large crescent cleft, thus enabling their access to inward-facing peptidyl-prolyl cis/trans isomerase catalytic sites and ipsilateral chaperone domain surfaces. As these chaperone surfaces participate extensively in crystal lattice contacts, we speculate that the observed lattice contacts reflect a proclivity for protein associations and represent substrate interactions by FKBP26 chaperone domains. Finally, we find that FKBP26 is an exceptionally flexible molecule, suggesting a mechanism for nonspecific substrate recognition.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA.