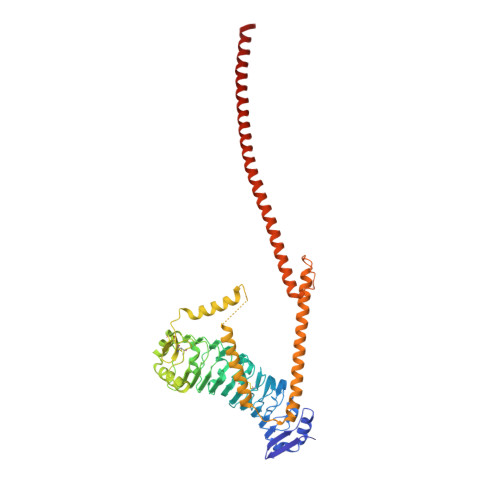
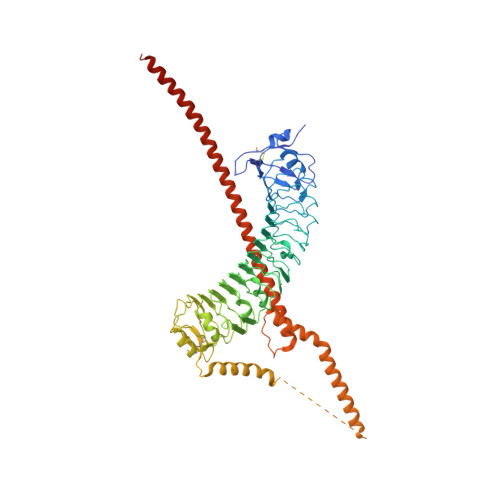
A heterodimeric complex of the LRR proteins LRIM1 and APL1C regulates complement-like immunity in Anopheles gambiae.
Baxter, R.H., Steinert, S., Chelliah, Y., Volohonsky, G., Levashina, E.A., Deisenhofer, J.(2010) Proc Natl Acad Sci U S A 107: 16817-16822
- PubMed: 20826443
- DOI: https://doi.org/10.1073/pnas.1010575107
- Primary Citation of Related Structures:
3O53, 3O6N, 3OJA - PubMed Abstract:
The leucine-rich repeat (LRR) proteins LRIM1 and APL1C control the function of the complement-like protein TEP1 in Anopheles mosquitoes. The molecular structure of LRIM1 and APL1C and the basis of their interaction with TEP1 represent a new type of innate immune complex. The LRIM1/APL1C complex specifically binds and solubilizes a cleaved form of TEP1 without an intact thioester bond. The LRIM1 and APL1C LRR domains have a large radius of curvature, glycosylated concave face, and a novel C-terminal capping motif. The LRIM1/APL1C complex is a heterodimer with a single intermolecular disulfide bond. The structure of the LRIM1/APL1C heterodimer reveals an interface between the two LRR domains and an extensive C-terminal coiled-coil domain. We propose that a cleaved form of TEP1 may act as a convertase for activation of other TEP1 molecules and that the LRIM1/APL1C heterodimer regulates formation of this TEP1 convertase.
Organizational Affiliation:
Howard Hughes Medical Institute and Department of Biochemistry, University of Texas Southwestern Medical Center, 6001 Forest Park Road, Dallas, TX 75390-9050, USA.