Structural Determinants for Activity and Specificity of the Bacterial Toxin LlpA
Ghequire, M.G., Garcia-Pino, A., Lebbe, E.K., Spaepen, S., Loris, R., De Mot, R.(2013) PLoS Pathog 9: e1003199-e1003199
- PubMed: 23468636
- DOI: https://doi.org/10.1371/journal.ppat.1003199
- Primary Citation of Related Structures:
3M7H, 3M7J, 4GC1, 4GC2 - PubMed Abstract:
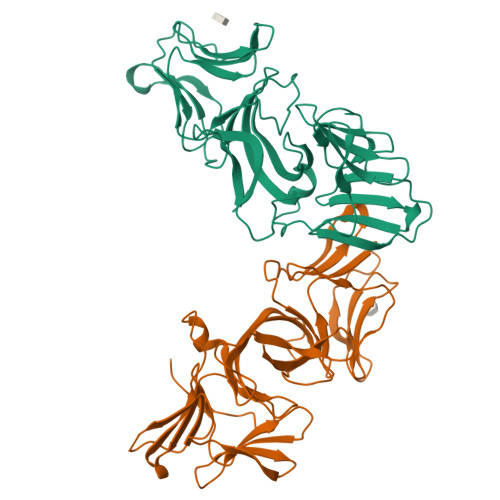
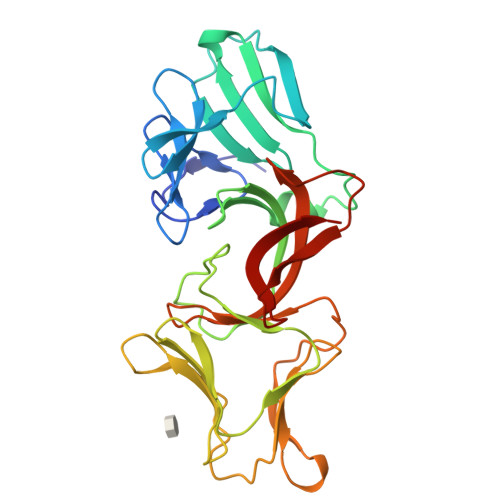
Lectin-like bacteriotoxic proteins, identified in several plant-associated bacteria, are able to selectively kill closely related species, including several phytopathogens, such as Pseudomonas syringae and Xanthomonas species, but so far their mode of action remains unrevealed. The crystal structure of LlpABW, the prototype lectin-like bacteriocin from Pseudomonas putida, reveals an architecture of two monocot mannose-binding lectin (MMBL) domains and a C-terminal β-hairpin extension. The C-terminal MMBL domain (C-domain) adopts a fold very similar to MMBL domains from plant lectins and contains a binding site for mannose and oligomannosides. Mutational analysis indicates that an intact sugar-binding pocket in this domain is crucial for bactericidal activity. The N-terminal MMBL domain (N-domain) adopts the same fold but is structurally more divergent and lacks a functional mannose-binding site. Differential activity of engineered N/C-domain chimers derived from two LlpA homologues with different killing spectra, disclosed that the N-domain determines target specificity. Apparently this bacteriocin is assembled from two structurally similar domains that evolved separately towards dedicated functions in target recognition and bacteriotoxicity.
Organizational Affiliation:
Centre of Microbial and Plant Genetics, University of Leuven, Heverlee-Leuven, Belgium.