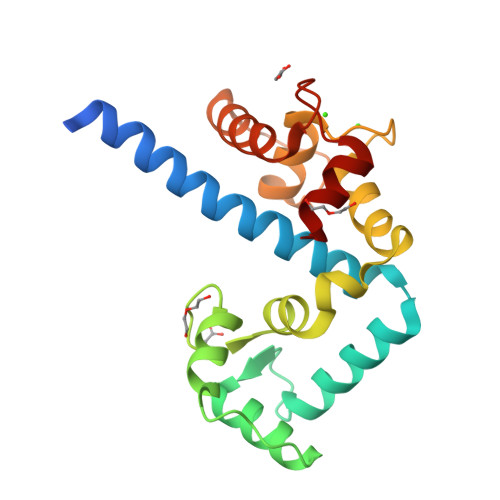
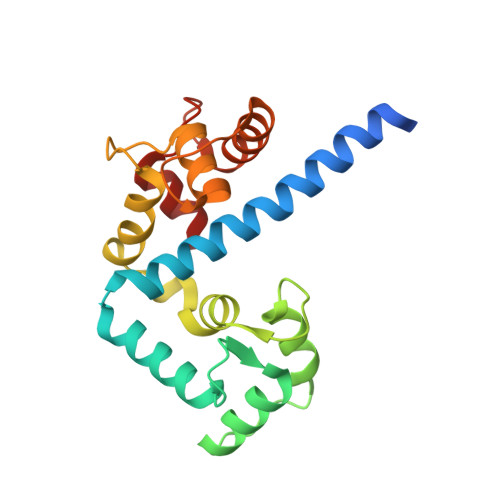
Structures of parasitic CDPK domains point to a common mechanism of activation.
Wernimont, A.K., Amani, M., Qiu, W., Pizarro, J.C., Artz, J.D., Lin, Y.H., Lew, J., Hutchinson, A., Hui, R.(2011) Proteins 79: 803-820
- PubMed: 21287613
- DOI: https://doi.org/10.1002/prot.22919
- Primary Citation of Related Structures:
3K21, 3KHE - PubMed Abstract:
We recently determined the first structures of inactivated and calcium-activated calcium-dependent protein kinases (CDPKs) from Apicomplexa. Calcium binding triggered a large conformational change that constituted a new mechanism in calcium signaling and a novel EF-hand fold (CAD, for CDPK activation domain). Thus we set out to determine if this mechanism was universal to all CDPKs. We solved additional CDPK structures, including one from the species Plasmodium. We highlight the similarities in sequence and structure across apicomplexan and plant CDPKs, and strengthen our observations that this novel mechanism could be universal to canonical CDPKs. Our new structures demonstrate more detailed steps in the mechanism of calcium activation and possible key players in regulation. Residues involved in making the largest conformational change are the most conserved across Apicomplexa, leading us to propose that the mechanism is indeed conserved. CpCDPK3_CAD and PfCDPK_CAD were captured at a possible intermediate conformation, lending insight into the order of activation steps. PfCDPK3_CAD adopts an activated fold, despite having an inactive EF-hand sequence in the N-terminal lobe. We propose that for most apicomplexan CDPKs, the mode of activation will be similar to that seen in our structures, while specific regulation of the inactive and active forms will require further investigation.
Organizational Affiliation:
Structural Genomics Consortium, University of Toronto, Toronto, Ontario, Canada M5G 1L7. amy.wernimont@utoronto.ca