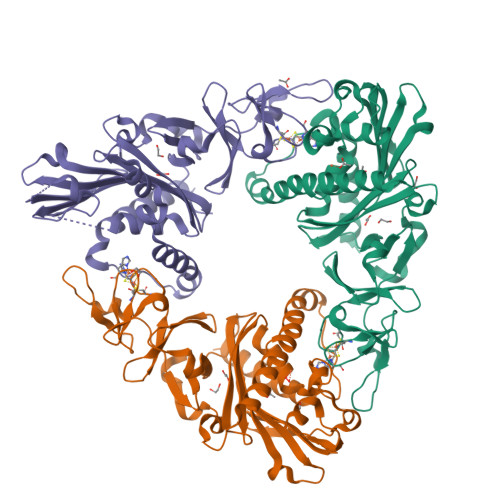
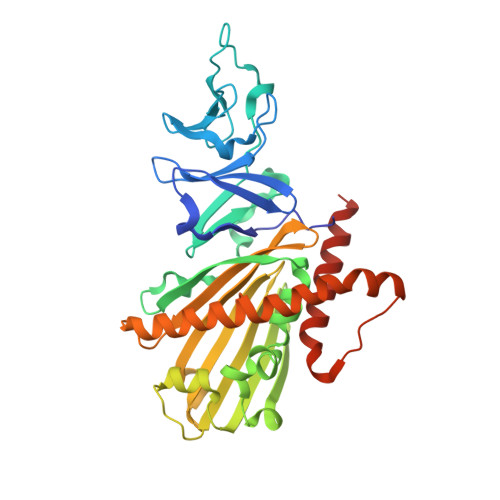
Crystal structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation.
Dumitru, R., Jiang, W.Z., Weeks, D.P., Wilson, M.A.(2009) J Mol Biology 392: 498-510
- PubMed: 19616011
- DOI: https://doi.org/10.1016/j.jmb.2009.07.021
- Primary Citation of Related Structures:
3GKE, 3GL0, 3GL2 - PubMed Abstract:
Dicamba (3,6-dichloro-2-methoxybenzoic acid) is a widely used herbicide that is efficiently degraded by soil microbes. These microbes use a novel Rieske nonheme oxygenase, dicamba monooxygenase (DMO), to catalyze the oxidative demethylation of dicamba to 3,6-dichlorosalicylic acid (DCSA) and formaldehyde. We have determined the crystal structures of DMO in the free state, bound to its substrate dicamba, and bound to the product DCSA at 2.10-1.75 A resolution. The structures show that the DMO active site uses a combination of extensive hydrogen bonding and steric interactions to correctly orient chlorinated, ortho-substituted benzoic-acid-like substrates for catalysis. Unlike other Rieske aromatic oxygenases, DMO oxygenates the exocyclic methyl group, rather than the aromatic ring, of its substrate. This first crystal structure of a Rieske demethylase shows that the Rieske oxygenase structural scaffold can be co-opted to perform varied types of reactions on xenobiotic substrates.
Organizational Affiliation:
Department of Biochemistry, The University of Nebraska-Lincoln, Lincoln, NE 68588-0664, USA.