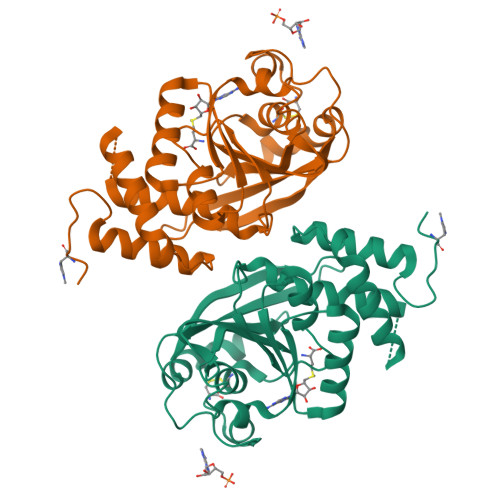
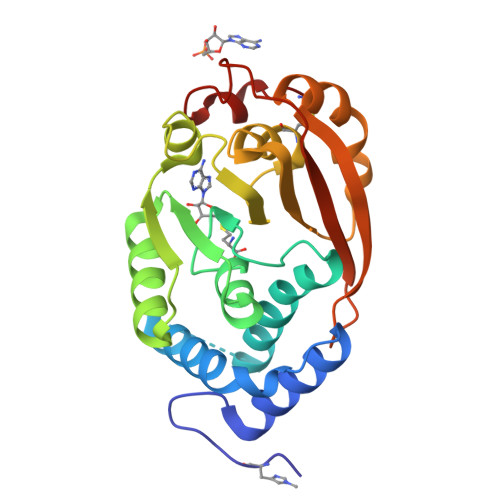
Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Gregory, S.T., Demirci, H., Belardinelli, R., Monshupanee, T., Gualerzi, C., Dahlberg, A.E., Jogl, G.(2009) RNA 15: 1693-1704
- PubMed: 19622680
- DOI: https://doi.org/10.1261/rna.1652709
- Primary Citation of Related Structures:
3G88, 3G89, 3G8A, 3G8B - PubMed Abstract:
The RsmG methyltransferase is responsible for N(7) methylation of G527 of 16S rRNA in bacteria. Here, we report the identification of the Thermus thermophilus rsmG gene, the isolation of rsmG mutants, and the solution of RsmG X-ray crystal structures at up to 1.5 A resolution. Like their counterparts in other species, T. thermophilus rsmG mutants are weakly resistant to the aminoglycoside antibiotic streptomycin. Growth competition experiments indicate a physiological cost to loss of RsmG activity, consistent with the conservation of the modification site in the decoding region of the ribosome. In contrast to Escherichia coli RsmG, which has been reported to recognize only intact 30S subunits, T. thermophilus RsmG shows no in vitro methylation activity against native 30S subunits, only low activity with 30S subunits at low magnesium concentration, and maximum activity with deproteinized 16S rRNA. Cofactor-bound crystal structures of RsmG reveal a positively charged surface area remote from the active site that binds an adenosine monophosphate molecule. We conclude that an early assembly intermediate is the most likely candidate for the biological substrate of RsmG.
Organizational Affiliation:
Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, Rhode Island 02912, USA.