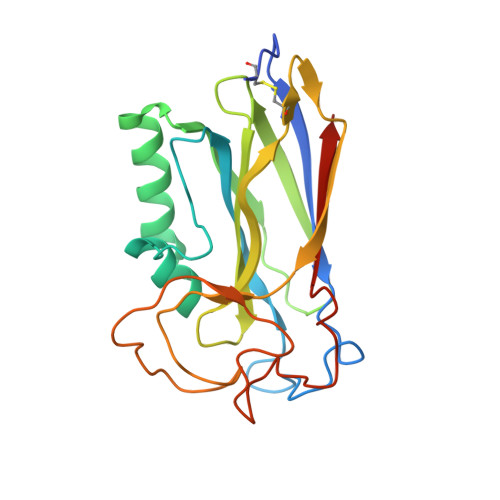
Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule.
Li, Y., Cao, C., Jia, W., Yu, L., Mo, M., Wang, Q., Huang, Y., Lim, J.M., Ishihara, M., Wells, L., Azadi, P., Robinson, H., He, Y.W., Zhang, L., Mariuzza, R.A.(2009) EMBO J 28: 286-297
- PubMed: 19153605
- DOI: https://doi.org/10.1038/emboj.2008.288
- Primary Citation of Related Structures:
3D34 - PubMed Abstract:
Mindin (spondin-2) is an extracellular matrix protein of unknown structure that is required for efficient T-cell priming by dendritic cells. Additionally, mindin functions as a pattern recognition molecule for initiating innate immune responses. These dual functions are mediated by interactions with integrins and microbial pathogens, respectively. Mindin comprises an N-terminal F-spondin (FS) domain and C-terminal thrombospondin type 1 repeat (TSR). We determined the structure of the FS domain at 1.8-A resolution. The structure revealed an eight-stranded antiparallel beta-sandwich motif resembling that of membrane-targeting C2 domains, including a bound calcium ion. We demonstrated that the FS domain mediates integrin binding and identified the binding site by mutagenesis. The mindin FS domain therefore represents a new integrin ligand. We further showed that mindin recognizes lipopolysaccharide (LPS) through its TSR domain, and obtained evidence that C-mannosylation of the TSR influences LPS binding. Through these dual interactions, the FS and TSR domains of mindin promote activation of both adaptive and innate immune responses.
Organizational Affiliation:
WM Keck Laboratory for Structural Biology, Center for Advanced Research in Biotechnology, University of Maryland Biotechnology Institute, Rockville, MD 20850, USA.