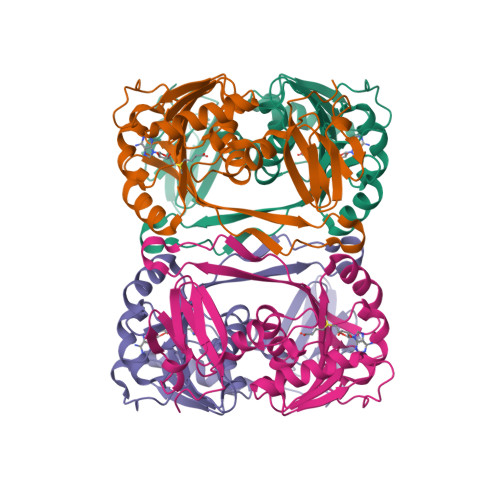
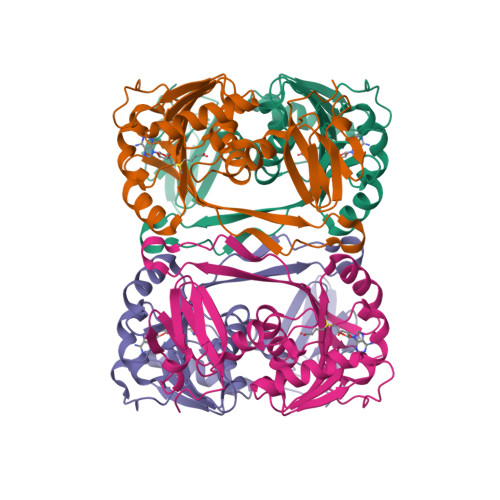
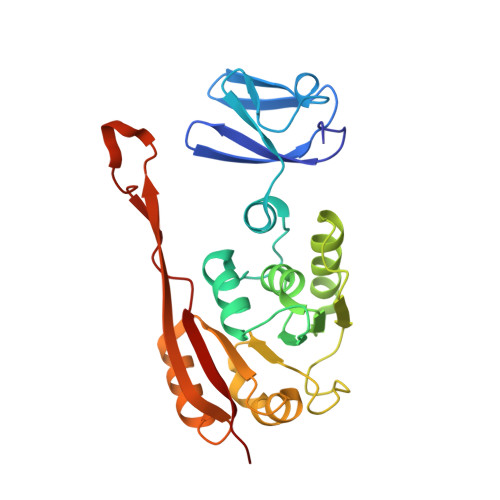
Crystal structure of tRNA m(1)A58 methyltransferase TrmI from Aquifex aeolicus in complex with S-adenosyl-L-methionine.
Kuratani, M., Yanagisawa, T., Ishii, R., Matsuno, M., Si, S.Y., Katsura, K., Ushikoshi-Nakayama, R., Shibata, R., Shirouzu, M., Bessho, Y., Yokoyama, S.(2014) J Struct Funct Genomics 15: 173-180
- PubMed: 24894648
- DOI: https://doi.org/10.1007/s10969-014-9183-0
- Primary Citation of Related Structures:
2YVL - PubMed Abstract:
The N (1)-methyladenosine residue at position 58 of tRNA is found in the three domains of life, and contributes to the stability of the three-dimensional L-shaped tRNA structure. In thermophilic bacteria, this modification is important for thermal adaptation, and is catalyzed by the tRNA m(1)A58 methyltransferase TrmI, using S-adenosyl-L-methionine (AdoMet) as the methyl donor. We present the 2.2 Å crystal structure of TrmI from the extremely thermophilic bacterium Aquifex aeolicus, in complex with AdoMet. There are four molecules per asymmetric unit, and they form a tetramer. Based on a comparison of the AdoMet binding mode of A. aeolicus TrmI to those of the Thermus thermophilus and Pyrococcus abyssi TrmIs, we discuss their similarities and differences. Although the binding modes to the N6 amino group of the adenine moiety of AdoMet are similar, using the side chains of acidic residues as well as hydrogen bonds, the positions of the amino acid residues involved in binding are diverse among the TrmIs from A. aeolicus, T. thermophilus, and P. abyssi.
Organizational Affiliation:
RIKEN Genomic Sciences Center, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, 230-0045, Japan.