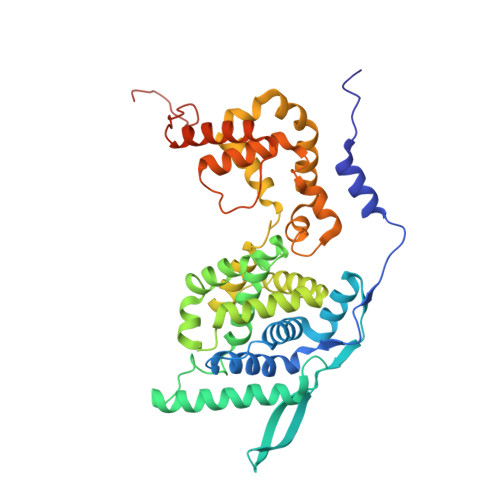
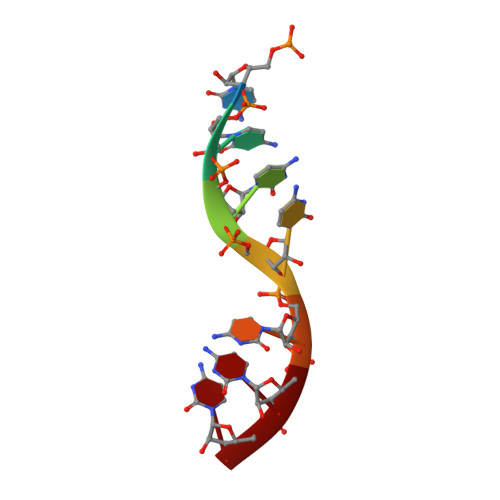
Crystal Structure of a Nucleocapsid-Like Nucleoprotein-RNA Complex of Respiratory Syncytial Virus
Tawar, R.G., Duquerroy, S., Vonrhein, C., Varela, P.F., Damier-Piolle, L., Castagne, N., Maclellan, K., Bedouelle, H., Bricogne, G., Bhella, D., Eleouet, J., Rey, F.A.(2009) Science 326: 1279
- PubMed: 19965480
- DOI: https://doi.org/10.1126/science.1177634
- Primary Citation of Related Structures:
2WJ8 - PubMed Abstract:
The respiratory syncytial virus (RSV) is an important human pathogen, yet neither a vaccine nor effective therapies are available to treat infection. To help elucidate the replication mechanism of this RNA virus, we determined the three-dimensional (3D) crystal structure at 3.3 A resolution of a decameric, annular ribonucleoprotein complex of the RSV nucleoprotein (N) bound to RNA. This complex mimics one turn of the viral helical nucleocapsid complex, which serves as template for viral RNA synthesis. The RNA wraps around the protein ring, with seven nucleotides contacting each N subunit, alternating rows of four and three stacked bases that are exposed and buried within a protein groove, respectively. Combined with electron microscopy data, this structure provides a detailed model for the RSV nucleocapsid, in which the bases are accessible for readout by the viral polymerase. Furthermore, the nucleoprotein structure highlights possible key sites for drug targeting.
Organizational Affiliation:
Institut Pasteur, Unité de Virologie Structurale, Département de Virologie and CNRS Unité de Recherche Associée (URA) 3015, 25 Rue du Dr Roux, 75724 Paris Cedex 15, France.