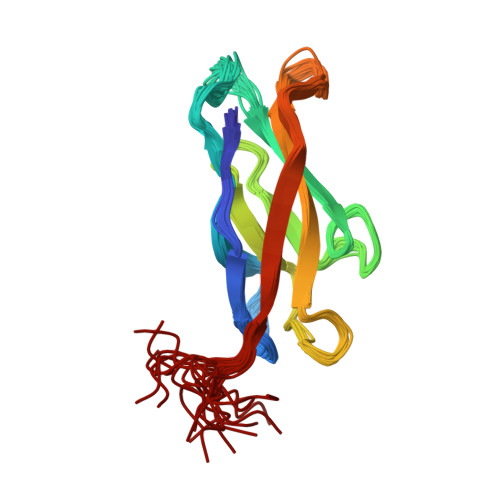
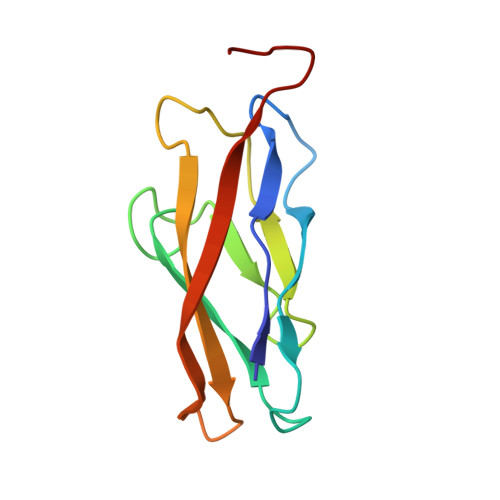
Insights into the low adhesive capacity of human T-cadherin from the NMR structure of Its N-terminal extracellular domain.
Dames, S.A., Bang, E., Haussinger, D., Ahrens, T., Engel, J., Grzesiek, S.(2008) J Biological Chem 283: 23485-23495
- PubMed: 18550521
- DOI: https://doi.org/10.1074/jbc.M708335200
- Primary Citation of Related Structures:
2V37 - PubMed Abstract:
T-cadherin is unique among the family of type I cadherins, because it lacks transmembrane and cytosolic domains, and attaches to the membrane via a glycophosphoinositol anchor. The N-terminal cadherin repeat of T-cadherin (Tcad1) is approximately 30% identical to E-, N-, and other classical cadherins. However, it lacks many amino acids crucial for their adhesive function of classical cadherins. Among others, Trp-2, which is the key residue forming the canonical strand-exchange dimer, is replaced by an isoleucine. Here, we report the NMR structure of the first cadherin repeat of T-cadherin (Tcad1). Tcad1, as other cadherin domains, adopts a beta-barrel structure with a Greek key folding topology. However, Tcad1 is monomeric in the absence and presence of calcium. Accordingly, lle-2 binds into a hydrophobic pocket on the same protomer and participates in an N-terminal beta-sheet. Specific amino acid replacements compared to classical cadherins reduce the size of the binding pocket for residue 2 and alter the backbone conformation and flexibility around residues 5 and 15 as well as many electrostatic interactions. These modifications apparently stabilize the monomeric form and make it less susceptible to a conformational switch upon calcium binding. The absence of a tendency for homoassociation observed by NMR is consistent with electron microscopy and solid-phase binding data of the full T-cadherin ectodomain (Tcad1-5). The apparent low adhesiveness of T-cadherin suggests that it is likely to be involved in reversible and dynamic cellular adhesion-deadhesion processes, which are consistent with its role in cell growth and migration.
Organizational Affiliation:
Department of Structural Biology, University of Basel, Klingelbergstr. 70, 4056 Basel, Switzerland. sonja.dames@unibas.ch