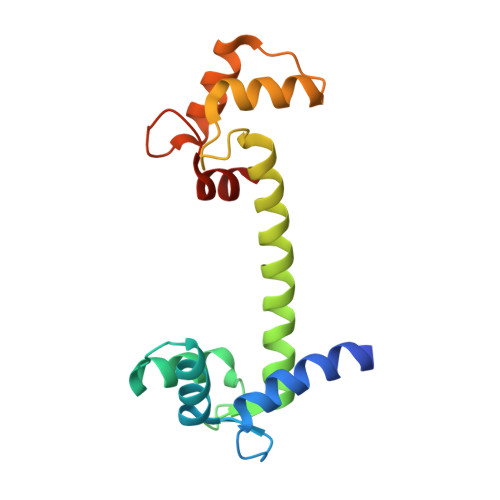
A Structural Insight Into Lead Neurotoxicity and Calmodulin Activation by Heavy Metals.
Kursula, P., Majava, V.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 653
- PubMed: 17671360
- DOI: https://doi.org/10.1107/S1744309107034525
- Primary Citation of Related Structures:
2V01, 2V02 - PubMed Abstract:
Calmodulin is a calcium sensor that is also capable of binding and being activated by other metal ions. Of specific interest in this respect is lead, which is known to be neurotoxic and to have a very high affinity towards calmodulin. Crystal structures of human calmodulin complexed with lead and barium ions have been solved. The results will help in understanding the activation mechanisms of calmodulin by different heavy metals and will provide a detailed view of a putative target for lead neurotoxicity in humans.
- Department of Biochemistry, University of Oulu, Oulu, Finland. petri.kursula@oulu.fi
Organizational Affiliation: