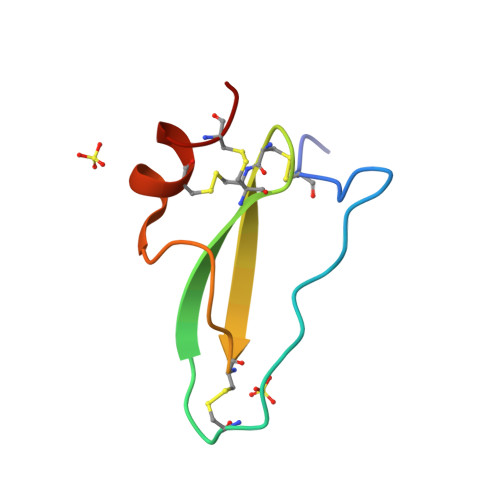
A Tick Protein with a Modified Kunitz Fold Inhibits Human Tryptase.
Paesen, G.C., Siebold, C., Harlos, K., Peacey, M.F., Nuttall, P.A., Stuart, D.I.(2007) J Mol Biology 368: 1172
- PubMed: 17391695
- DOI: https://doi.org/10.1016/j.jmb.2007.03.011
- Primary Citation of Related Structures:
2UUX, 2UUY - PubMed Abstract:
TdPI, a tick salivary gland product related to Kunitz/BPTI proteins is a potent inhibitor of human beta-tryptase. Kinetic assays suggest that three of the four catalytic sites of tryptase are blocked by TdPI, and that the inhibition of one of these involves a peptide flanking the Kunitz head. In the course of the inhibition, tryptase cleaves TdPI at several positions. Crystal structures of the TdPI head, on its own and in complex with trypsin, reveal features that are not found in classical Kunitz/BPTI proteins and suggest the mode of interaction with tryptase. The loop of TdPI connecting the beta-sheet with the C-terminal alpha-helix is shortened, the disulphide-bridge pattern altered and N and C termini separated to produce a highly pointed molecule capable of penetrating the cramped active sites of tryptase. TdPI accumulates in the cytosolic granules of mast cells, presumably suppressing inflammation in the host animal's skin by tryptase inhibition.
Organizational Affiliation:
CEH, Mansfield Road, Oxford, OX1 3SR, UK. gcp@ceh.ac.uk