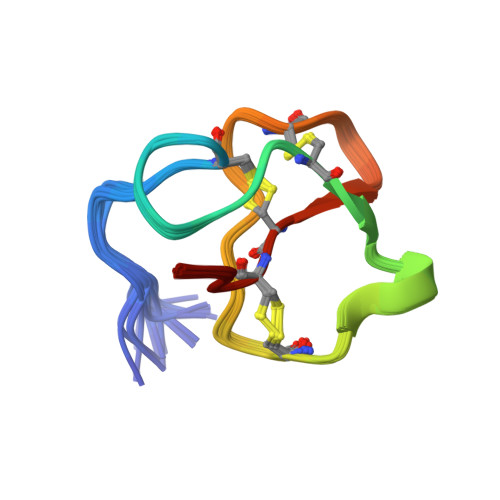
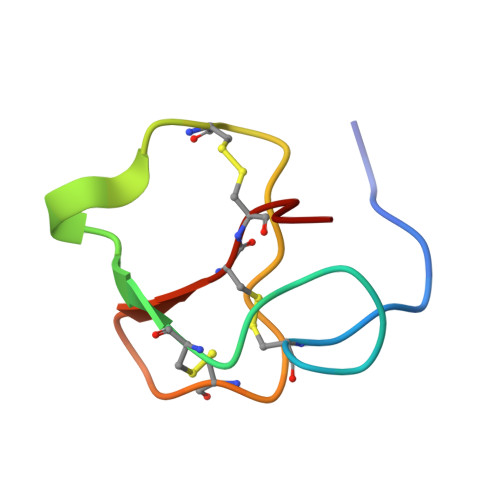
Structure, self-assembly, and dual role of a beta-defensin-like peptide from the Chinese soft-shelled turtle eggshell matrix.
Lakshminarayanan, R., Vivekanandan, S., Samy, R.P., Banerjee, Y., Chi-Jin, E.O., Teo, K.W., Jois, S.D., Kini, R.M., Valiyaveettil, S.(2008) J Am Chem Soc 130: 4660-4668
- PubMed: 18341335
- DOI: https://doi.org/10.1021/ja075659k
- Primary Citation of Related Structures:
2JR3 - PubMed Abstract:
Biomineral matrix formation and molecular recognition are two important processes associated with eggshell biomineralization. To understand these two processes, a major intracrystalline peptide, pelovaterin, was isolated from turtle (Pelodiscus sinensis) eggshell and its tertiary and quaternary structures were established. The global fold of pelovaterin is similar to that of human beta-defensins but has a large hydrophobic core and a short hydrophilic N-terminal segment, which is not preserved in defensins. Pelovaterin exhibits strong antimicrobial activity against two pathogenic gram-negative bacteria, Pseudomonas aeruginosa and Proteus vulgaris, and stabilizes a thin film of metastable vaterite. We show that pelovaterin self-aggregates in the form of micellar nanospheres and the aggregation in solution is entropy-driven. It is suggested that the micellar aggregation of pelovaterin is responsible for the induction and stabilization of the metastable phase by altering the interfacial energy. The results demonstrate the adaptability of an extracellular matrix protein to perform multiple tasks: polymorph discrimination and protection of the contents of the egg against bacterial invasion.
Organizational Affiliation:
Department of Chemistry, Faculty of Science, 3 Science Drive 3, National University of Singapore, Singapore 117543.