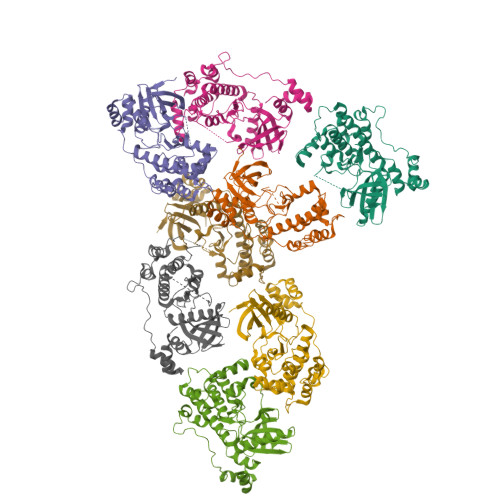
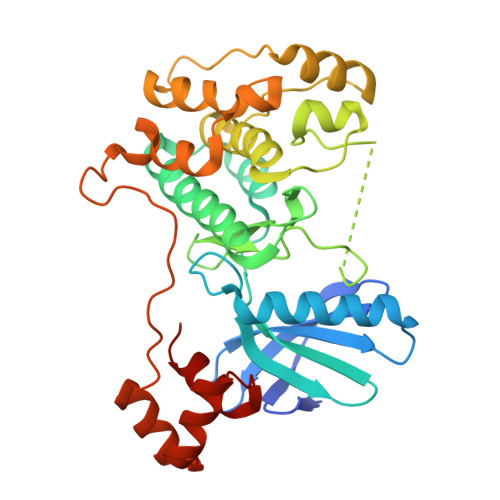
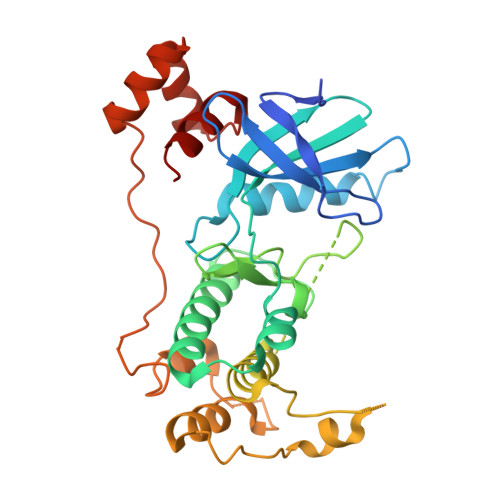
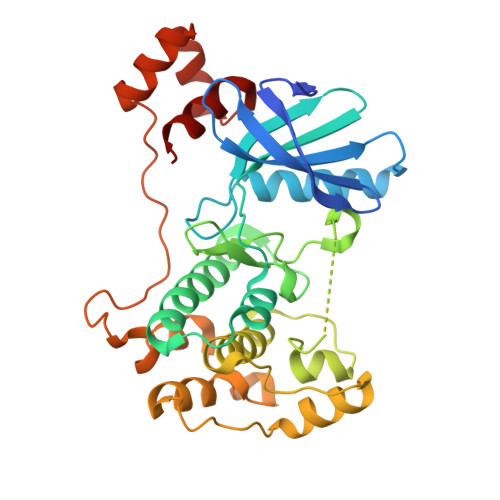
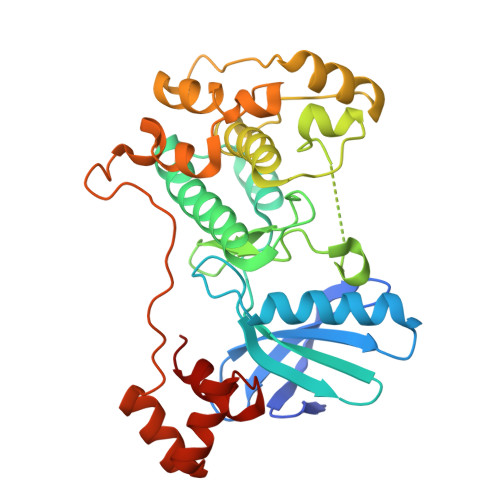
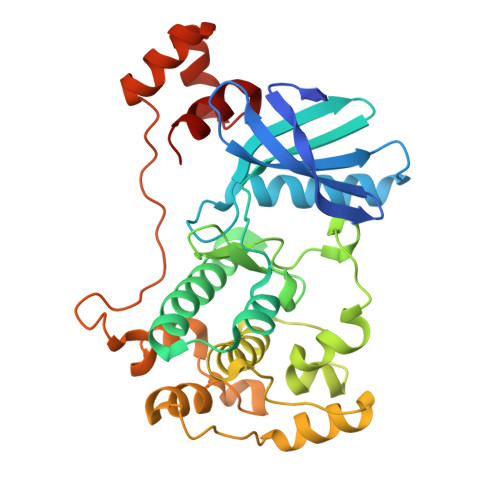
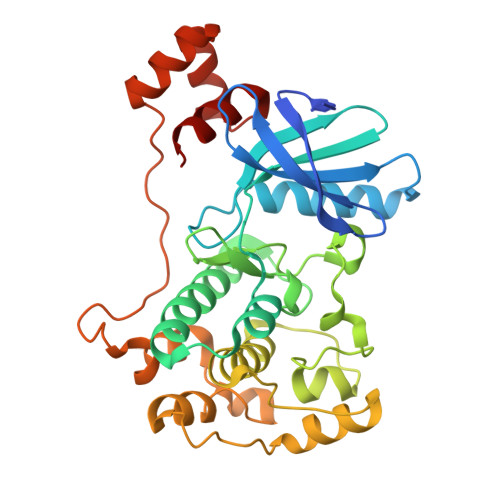
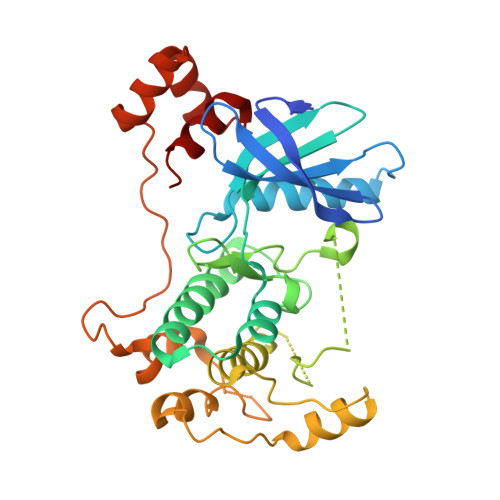
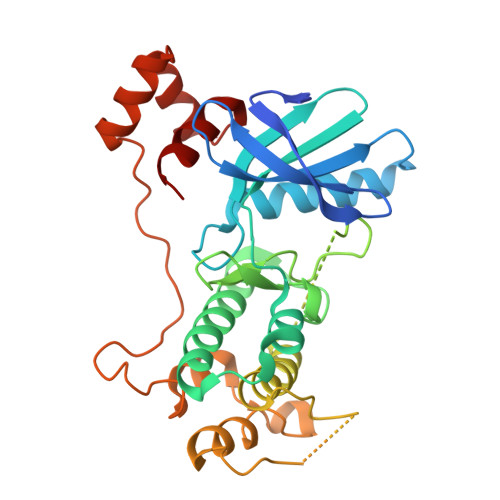
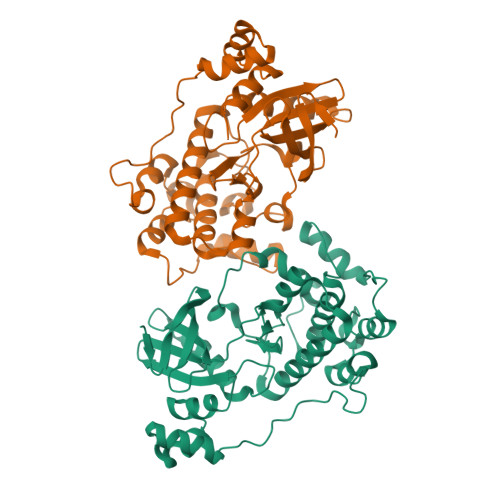
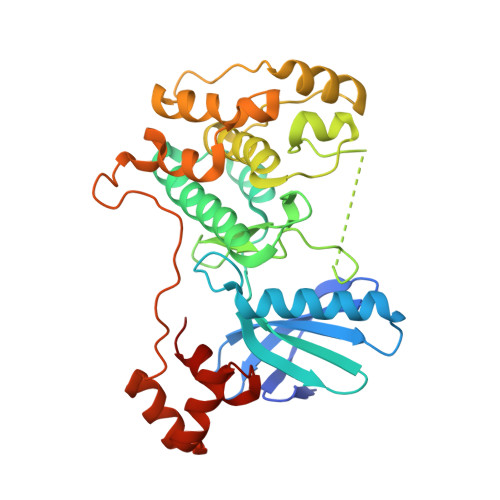
Structural variations in the catalytic and ubiquitin-associated domains of microtubule-associated protein/microtubule affinity regulating kinase (MARK) 1 and MARK2
Marx, A., Nugoor, C., Mueller, J., Panneerselvam, S., Timm, T., Bilang, M., Mylonas, E., Svergun, D.I., Mandelkow, E.-M., Mandelkow, E.(2006) J Biological Chem 281: 27586-27599
- PubMed: 16803889
- DOI: https://doi.org/10.1074/jbc.M604865200
- Primary Citation of Related Structures:
2HAK - PubMed Abstract:
The microtubule-associated protein (MAP)/microtubule affinity regulating kinase (MARK)/Par-1 phosphorylates microtubule-associated proteins tau, MAP2, and MAP4 and is involved in the regulation of microtubule-based transport. Par-1, a homologue of MARK in Drosophila and Caenorhabditis elegans, is essential for the development of embryonic polarity. Four isoforms of MARK are found in humans. Recently, we reported the crystal structure of the catalytic and ubiquitin-associated domains of MARK2, an isoform enriched in brain (Panneerselvam, S., Marx, A., Mandelkow, E.-M., and Mandelkow, E. (2006) Structure 14, 173-183). It showed that the ubiquitin-associated domain (UBA) domain has an unusual fold and binds to the N-terminal lobe of the catalytic domain. This is at variance with a previous low resolution structure derived from small angle solution scattering (Jaleel, M., Villa, F., Deak, M., Toth, R., Prescott, A. R., Van Aalten, D. M., and Alessi, D. R. (2006) Biochem. J. 394, 545-555), which predicts binding of the UBA domain to the larger, C-terminal lobe. Here we report the crystal structure of the catalytic and UBA domain of another isoform, MARK1. Although the crystal packing of the two isoforms are unrelated, the overall conformations of the molecules are similar. Notably, the UBA domain has the same unusual conformation as in MARK2, and it binds at the same site. Remarkable differences occur in the catalytic domain at helix C, the catalytic loop, and the activation segment.
Organizational Affiliation:
Max-Planck-Unit for Structural Molecular Biology, Notkestrasse 85, 22607 Hamburg, Germany.