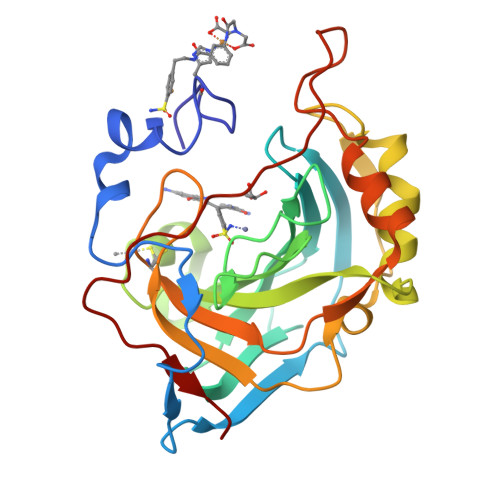
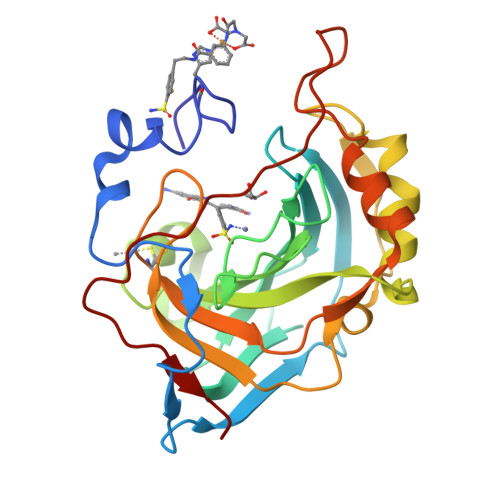
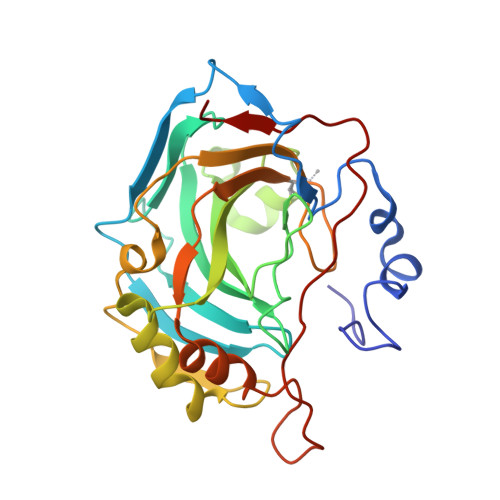
Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with two-prong inhibitors reveal the molecular basis of high affinity.
Jude, K.M., Banerjee, A.L., Haldar, M.K., Manokaran, S., Roy, B., Mallik, S., Srivastava, D.K., Christianson, D.W.(2006) J Am Chem Soc 128: 3011-3018
- PubMed: 16506782
- DOI: https://doi.org/10.1021/ja057257n
- Primary Citation of Related Structures:
2FOQ, 2FOS, 2FOU, 2FOV, 2FOY - PubMed Abstract:
The atomic-resolution crystal structures of human carbonic anhydrases I and II complexed with "two-prong" inhibitors are reported. Each inhibitor contains a benzenesulfonamide prong and a cupric iminodiacetate (IDA-Cu(2+)) prong separated by linkers of different lengths and compositions. The ionized NH(-) group of each benzenesulfonamide coordinates to the active site Zn(2+) ion; the IDA-Cu(2+) prong of the tightest-binding inhibitor, BR30, binds to H64 of CAII and H200 of CAI. This work provides the first evidence verifying the structural basis of nanomolar affinity measured for two-prong inhibitors targeting the carbonic anhydrases.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104, USA.