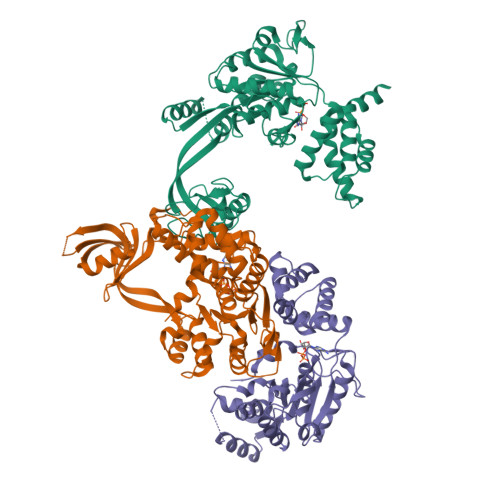
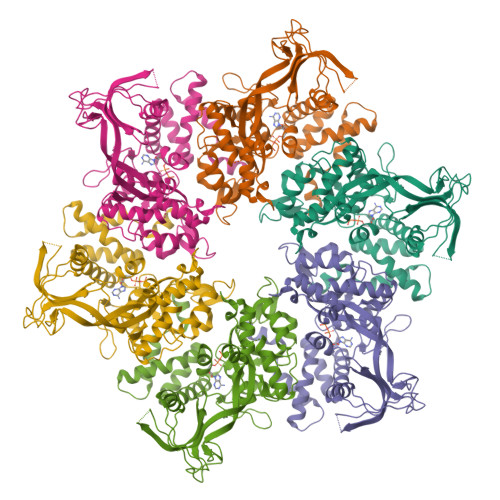
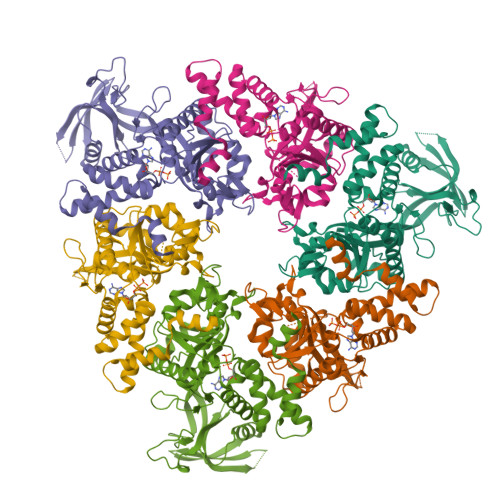
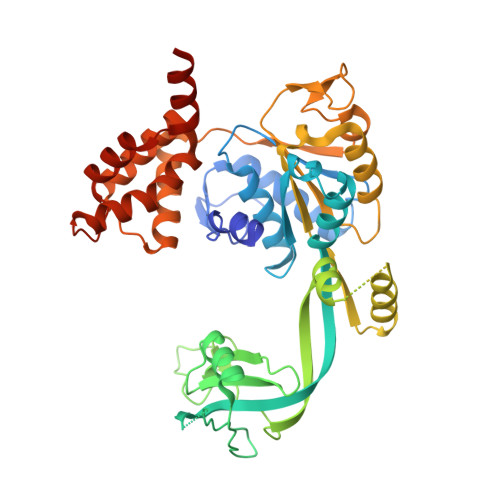
Crystal structure of the human AAA+ protein RuvBL1.
Matias, P.M., Gorynia, S., Donner, P., Carrondo, M.A.(2006) J Biological Chem 281: 38918-38929
- PubMed: 17060327
- DOI: https://doi.org/10.1074/jbc.M605625200
- Primary Citation of Related Structures:
2C9O - PubMed Abstract:
RuvBL1 is an evolutionarily highly conserved eukaryotic protein belonging to the AAA(+)-family of ATPases (ATPase associated with diverse cellular activities). It plays important roles in essential signaling pathways such as the c-Myc and Wnt pathways in chromatin remodeling, transcriptional and developmental regulation, and DNA repair and apoptosis. Herein we present the three-dimensional structure of the selenomethionine variant of human RuvBL1 refined using diffraction data to 2.2A of resolution. The crystal structure of the hexamer is formed of ADP-bound RuvBL1 monomers. The monomers contain three domains, of which the first and the third are involved in ATP binding and hydrolysis. Although it has been shown that ATPase activity of RuvBL1 is needed for several in vivo functions, we could only detect a marginal activity with the purified protein. Structural homology and DNA binding studies demonstrate that the second domain, which is unique among AAA(+) proteins and not present in the bacterial homolog RuvB, is a novel DNA/RNA-binding domain. We were able to demonstrate that RuvBL1 interacted with single-stranded DNA/RNA and double-stranded DNA. The structure of the RuvBL1.ADP complex, combined with our biochemical results, suggest that although RuvBL1 has all the structural characteristics of a molecular motor, even of an ATP-driven helicase, one or more as yet undetermined cofactors are needed for its enzymatic activity.
Organizational Affiliation:
Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Apartado 127, 2781-901 Oeiras, Portugal.