Plasminogen activator inhibitor-2 is highly tolerant to P8 residue substitution--implications for serpin mechanistic model and prediction of nsSNP activities
Di Giusto, D.A., Sutherland, A.P., Jankova, L., Harrop, S.J., Curmi, P.M., King, G.C.(2005) J Mol Biology 353: 1069-1080
- PubMed: 16214170
- DOI: https://doi.org/10.1016/j.jmb.2005.09.008
- Primary Citation of Related Structures:
2ARQ, 2ARR - PubMed Abstract:
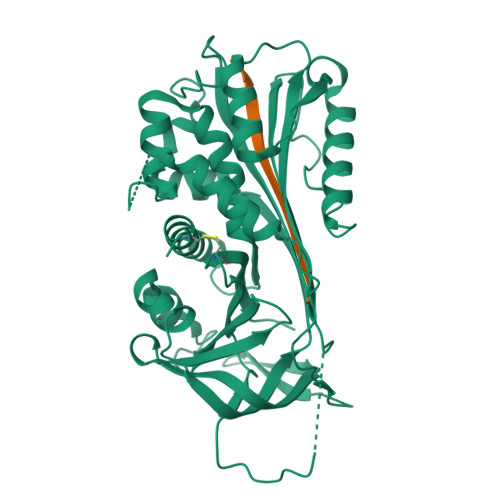
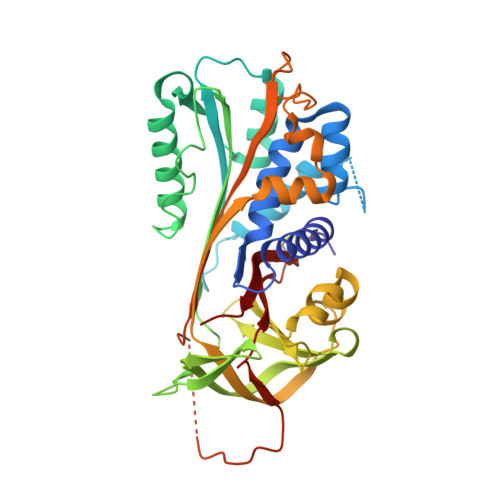
The serine protease inhibitor (serpin) superfamily is involved in a wide range of cellular processes including fibrinolysis, angiogenesis, apoptosis, inflammation, metastasis and viral pathogenesis. Here, we investigate the unique mousetrap inhibition mechanism of serpins through saturation mutagenesis of the P8 residue for a typical family member, plasminogen activator inhibitor-2 (PAI-2). A number of studies have proposed an important role for the P8 residue in the efficient insertion and stabilisation of the cleaved reactive centre loop (RCL), which is a key event in the serpin inhibitory mechanism. The importance of this residue for inhibition of the PAI-2 protease target urinary plasminogen activator (urokinase, uPA) is confirmed, although a high degree of tolerance to P8 substitution is observed. Out of 19 possible PAI-2 P8 mutants, 16 display inhibitory activities within an order of magnitude of the wild-type P8 Thr species. Crystal structures of complexes between PAI-2 and RCL-mimicking peptides with P8 Met or Asp mutations are determined, and structural comparison with the wild-type complex substantiates the ability of the S8 pocket to accommodate disparate side-chains. These data indicate that the identity of the P8 residue is not a determinant of efficient RCL insertion, and provide further evidence for functional plasticity of key residues within enzyme structures. Poor correlation of observed PAI-2 P8 mutant activities with a range of physicochemical, evolutionary and thermodynamic predictive indices highlights the practical limitations of existing approaches to predicting the molecular phenotype of protein variants.
Organizational Affiliation:
School of Biotechnology and Biomolecular Sciences, The University of New South Wales, Sydney, NSW 2052, Australia.