Pros and cons of cryocrystallography: should we also collect a room-temperature data set?
Dunlop, K.V., Irvin, R.T., Hazes, B.(2005) Acta Crystallogr D Biol Crystallogr 61: 80-87
- PubMed: 15608379
- DOI: https://doi.org/10.1107/S0907444904027179
- Primary Citation of Related Structures:
1X6P, 1X6Q, 1X6R, 1X6X, 1X6Y, 1X6Z - PubMed Abstract:
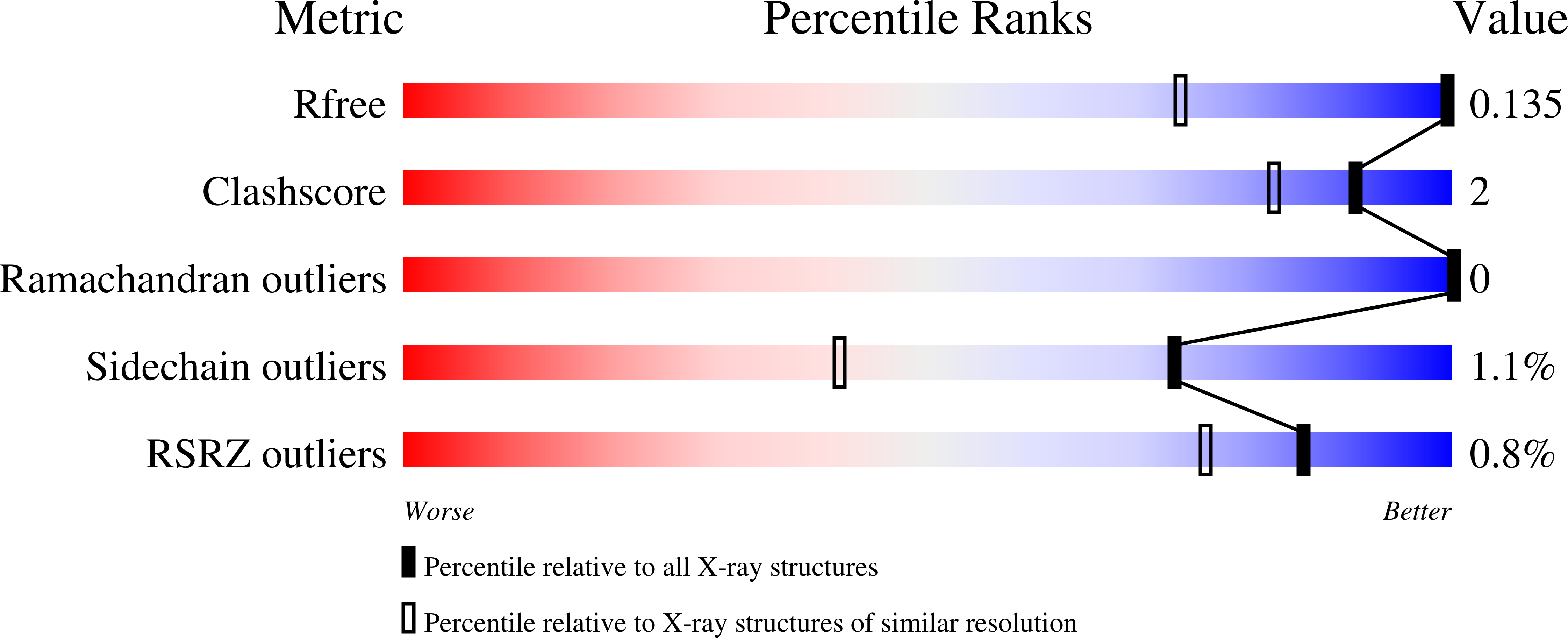
High-resolution protein structures are becoming more common owing to the availability of increasingly brilliant synchrotron X-ray sources. However, to withstand the increased X-ray dose the crystals must be held at cryogenic temperatures. To compare the benefit of increased resolution with the drawback of potential temperature-induced changes, three room-temperature and three cryogenic data sets for PAK pilin have been collected at resolutions between 1.8 and 0.78 A. The results show that although the high-resolution cryogenic structures are more precise and more detailed, they also show systematic deviations from the room-temperature structures. Small but significant differences are even observed in the structural core, whilst more extensive changes occur at the protein surface. These differences can affect biological interpretations, especially because many important biological processes take place at the protein surface. Accordingly, although high-quality cryogenic synchrotron data is extremely valuable to protein crystallography, room-temperature structures are still desirable, especially if the research question involves protein features that are sensitive to temperature-induced changes.
Organizational Affiliation:
Department of Medical Microbiology and Immunology, University of Alberta, Canada.