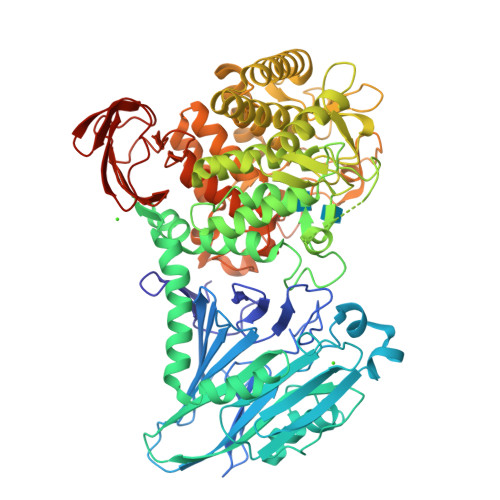
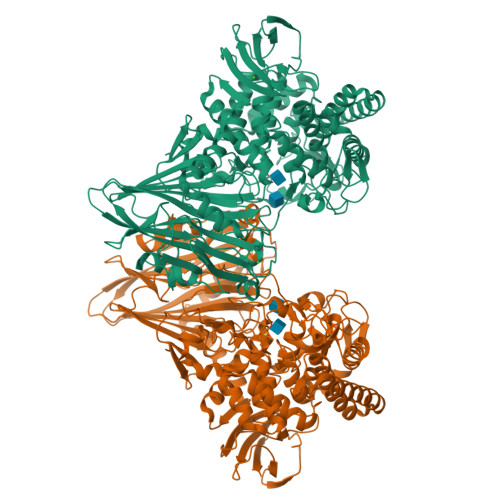
Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Hidaka, M., Honda, Y., Kitaoka, M., Nirasawa, S., Hayashi, K., Wakagi, T., Shoun, H., Fushinobu, S.(2004) Structure 12: 937-947
- PubMed: 15274915
- DOI: https://doi.org/10.1016/j.str.2004.03.027
- Primary Citation of Related Structures:
1V7V, 1V7W, 1V7X - PubMed Abstract:
Vibrio proteolyticus chitobiose phosphorylase (ChBP) belongs to glycosyl transferase family 36 (GT-36), and catalyzes the reversible phosphorolysis of chitobiose into alpha-GlcNAc-1-phosphate and GlcNAc with inversion of the anomeric configuration. As the first known structures of a GT-36 enzyme, we determined the crystal structure of ChBP in a ternary complex with GlcNAc and SO(4). It is also the first structures of an inverting phosphorolytic enzyme in a complex with a sugar and a sulfate ion, and reveals a pseudo-ternary complex structure of enzyme-sugar-phosphate. ChBP comprises a beta sandwich domain and an (alpha/alpha)(6) barrel domain, constituting a distinctive structure among GT families. Instead, it shows significant structural similarity with glycoside hydrolase (GH) enzymes, glucoamylases (GH-15), and maltose phosphorylase (GH-65) in clan GH-L. The structural similarity reported here, together with distant sequence similarities between ChBP and GHs, led to the reclassification of family GT-36 into a novel GH family, namely GH-94.
Organizational Affiliation:
Department of Biotechnology, The University of Tokyo, 1-1-1, Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan.