Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene.
Miller, D.J., Shuvalova, L., Evdokimova, E., Savchenko, A., Yakunin, A.F., Anderson, W.F.(2007) Protein Sci 16: 1338-1348
- PubMed: 17586769
- DOI: https://doi.org/10.1110/ps.072764907
- Primary Citation of Related Structures:
1SU1 - PubMed Abstract:
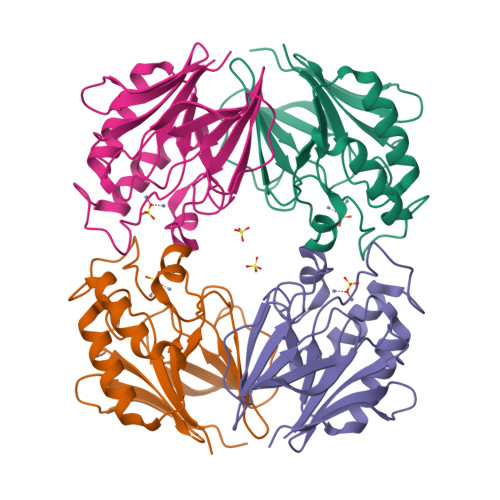
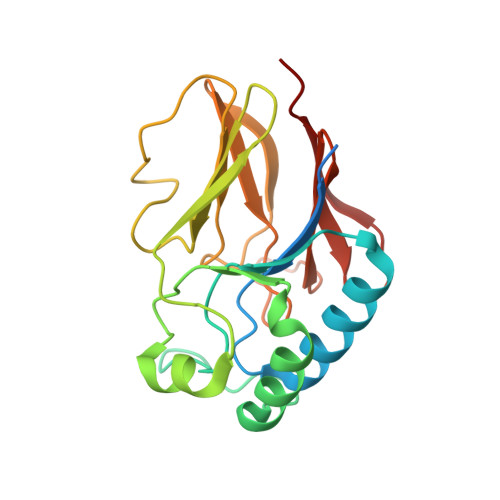
Escherichia coli YfcE belongs to a conserved protein family within the calcineurin-like phosphoesterase superfamily (Pfam00149) that is widely distributed in bacteria and archaea. Superfamily members are metallophosphatases that include monoesterases and diesterases involved in a variety of cellular functions. YfcE exhibited catalytic activity against bis-p-nitrophenyl phosphate, a general substrate for phosphodiesterases, and had an absolute requirement for Mn2+. However, no activity was observed with phosphodiesters and over 50 naturally occurring phosphomonoesters. The crystal structure of the YfcE phosphodiesterase has been determined to 2.25 A resolution. YfcE has a beta-sandwich architecture similar to metallophosphatases of common ancestral origin. Unlike its more complex homologs that have added structural elements for regulation and substrate recognition, the relatively small 184-amino-acid protein has retained its ancestral simplicity. The tetrameric protein carries two zinc ions per active site from the E. coli extract that reflect the conserved di-Mn2+ active site geometry. A cocrystallized sulfate inhibitor mimics the binding of phosphate moeities in known ligand/phosphatase complexes. Thus, YfcE has a similar active site and biochemical mechanism as well-characterized superfamily members, while the YfcE phosphodiester-containing substrate is unique.
Organizational Affiliation:
Department of Molecular Pharmacology and Biological Chemistry, Northwestern University, Feinberg School of Medicine, Chicago, IL 60611, USA.