Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
Saul, F.A., Arie, J.P., Vulliez-le Normand, B., Kahn, R., Betton, J.M., Bentley, G.A.(2004) J Mol Biology 335: 595-608
- PubMed: 14672666
- DOI: https://doi.org/10.1016/j.jmb.2003.10.056
- Primary Citation of Related Structures:
1Q6H, 1Q6I, 1Q6U - PubMed Abstract:
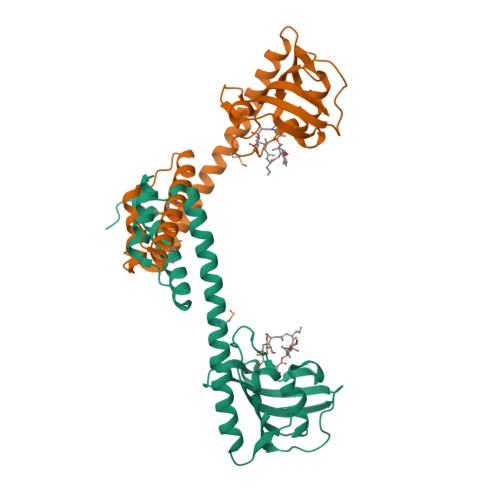
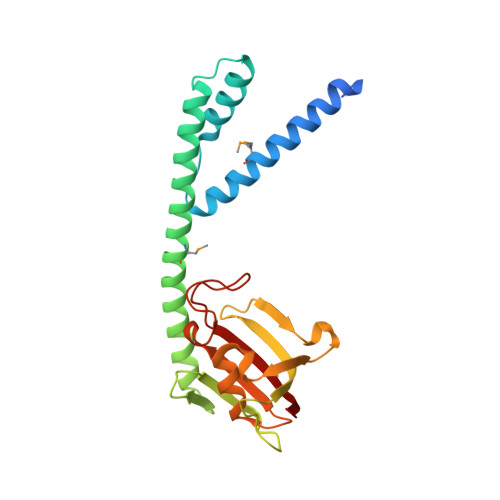
The protein FkpA from the periplasm of Escherichia coli exhibits both cis/trans peptidyl-prolyl isomerase (PPIase) and chaperone activities. The crystal structure of the protein has been determined in three different forms: as the full-length native molecule, as a truncated form lacking the last 21 residues, and as the same truncated form in complex with the immunosuppressant ligand, FK506. FkpA is a dimeric molecule in which the 245-residue subunit is divided into two domains. The N-terminal domain includes three helices that are interlaced with those of the other subunit to provide all inter-subunit contacts maintaining the dimeric species. The C-terminal domain, which belongs to the FK506-binding protein (FKBP) family, binds the FK506 ligand. The overall form of the dimer is V-shaped, and the different crystal structures reveal a flexibility in the relative orientation of the two C-terminal domains located at the extremities of the V. The deletion mutant FkpNL, comprising the N-terminal domain only, exists in solution as a mixture of monomeric and dimeric species, and exhibits chaperone activity. By contrast, a deletion mutant comprising the C-terminal domain only is monomeric, and although it shows PPIase activity, it is devoid of chaperone function. These results suggest that the chaperone and catalytic activities reside in the N and C-terminal domains, respectively. Accordingly, the observed mobility of the C-terminal domains of the dimeric molecule could effectively adapt these two independent folding functions of FkpA to polypeptide substrates.
Organizational Affiliation:
Unité d'Immunologie Structurale (C.N.R.S. U.R.A. 2185), Institut Pasteur, 25 rue du Dr Roux, 75724 Paris cedex 15, France.