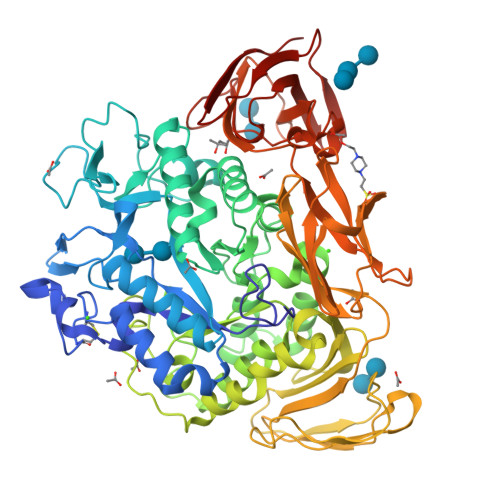
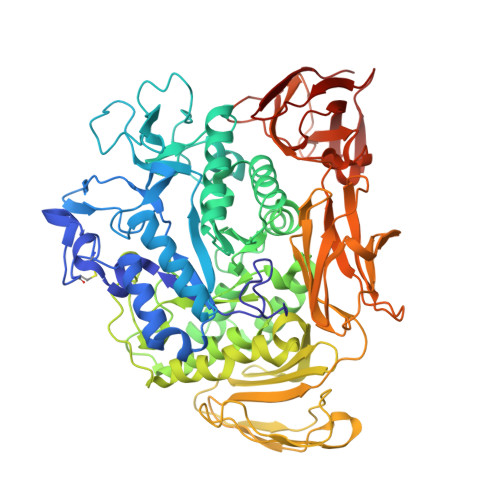
Conversion of Cyclodextrin Glycosyltransferase into a Starch Hydrolase by Directed Evolution: The Role of Alanine 230 in Acceptor Subsite +1
Leemhuis, H., Rozeboom, H.J., Wilbrink, M., Euverink, G.J., Dijkstra, B.W., Dijkhuizen, L.(2003) Biochemistry 42: 7518-7526
- PubMed: 12809508
- DOI: https://doi.org/10.1021/bi034439q
- Primary Citation of Related Structures:
1PEZ - PubMed Abstract:
Cyclodextrin glycosyltransferase (CGTase) preferably catalyzes transglycosylation reactions, whereas many other alpha-amylase family enzymes are hydrolases. Despite the availability of three-dimensional structures of several transglycosylases and hydrolases of this family, the factors that determine the hydrolysis and transglycosylation specificity are far from understood. To identify the amino acid residues that are critical for the transglycosylation reaction specificity, we carried out error-prone PCR mutagenesis and screened for Bacillus circulans strain 251 CGTase mutants with increased hydrolytic activity. After three rounds of mutagenesis the hydrolytic activity had increased 90-fold, reaching the highest hydrolytic activity ever reported for a CGTase. The single mutation with the largest effect (A230V) occurred in a residue not studied before. The structure of this A230V mutant suggests that the larger valine side chain hinders substrate binding at acceptor subsite +1, although not to the extent that catalysis is impossible. The much higher hydrolytic than transglycosylation activity of this mutant indicates that the use of sugar acceptors is hindered especially. This observation is in favor of a proposed induced-fit mechanism, in which sugar acceptor binding at acceptor subsite +1 activates the enzyme in transglycosylation [Uitdehaag et al. (2000) Biochemistry 39, 7772-7780]. As the A230V mutation introduces steric hindrance at subsite +1, this mutation is expected to negatively affect the use of sugar acceptors. Thus, the characteristics of mutant A230V strongly support the existence of the proposed induced-fit mechanism in which sugar acceptor binding activates CGTase in a transglycosylation reaction.
Organizational Affiliation:
Department of Microbiology, Groningen Biomolecular Sciences and Biotechnology Institute (GBB), University of Groningen, Kerklaan 30, 9751 NN Haren, The Netherlands.