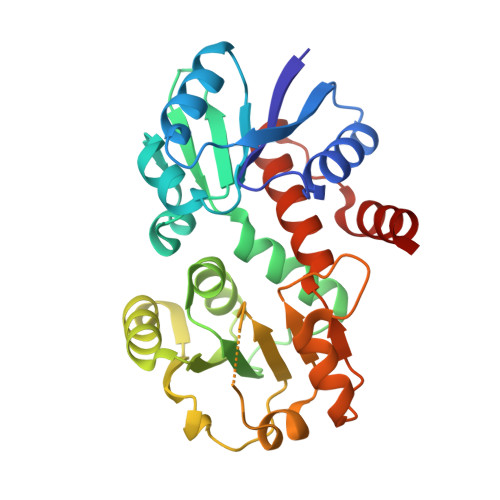
The crystal structure of shikimate dehydrogenase (AroE) reveals a unique NADPH binding mode
Ye, S., von Delft, F., Brooun, A., Knuth, M.W., Swanson, R.V., McRee, D.E.(2003) J Bacteriol 185: 4144-4151
- PubMed: 12837789
- DOI: https://doi.org/10.1128/JB.185.14.4144-4151.2003
- Primary Citation of Related Structures:
1P74, 1P77 - PubMed Abstract:
Shikimate dehydrogenase catalyzes the NADPH-dependent reversible reduction of 3-dehydroshikimate to shikimate. We report the first X-ray structure of shikimate dehydrogenase from Haemophilus influenzae to 2.4-A resolution and its complex with NADPH to 1.95-A resolution. The molecule contains two domains, a catalytic domain with a novel open twisted alpha/beta motif and an NADPH binding domain with a typical Rossmann fold. The enzyme contains a unique glycine-rich P-loop with a conserved sequence motif, GAGGXX, that results in NADPH adopting a nonstandard binding mode with the nicotinamide and ribose moieties disordered in the binary complex. A deep pocket with a narrow entrance between the two domains, containing strictly conserved residues primarily contributed by the catalytic domain, is identified as a potential 3-dehydroshikimate binding pocket. The flexibility of the nicotinamide mononucleotide portion of NADPH may be necessary for the substrate 3-dehydroshikimate to enter the pocket and for the release of the product shikimate.
Organizational Affiliation:
Syrrx Inc., San Diego, California 92121, USA.