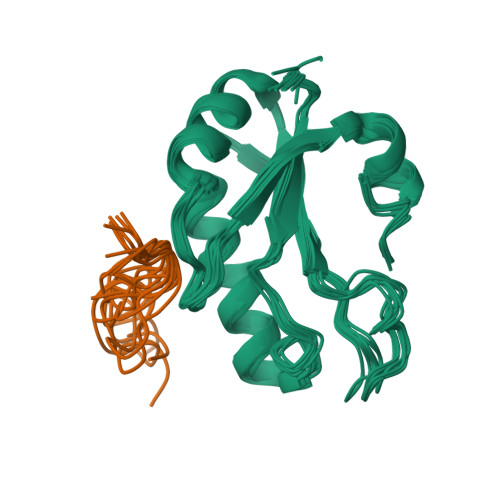
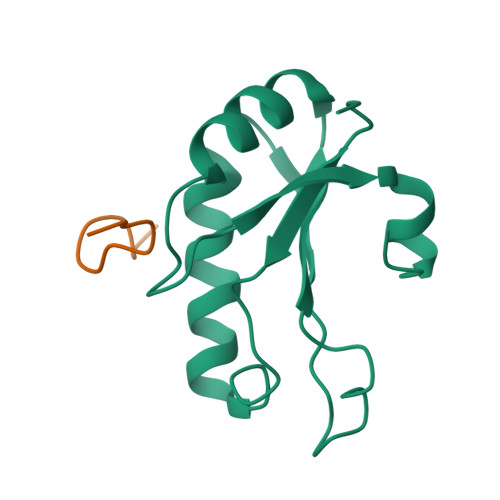
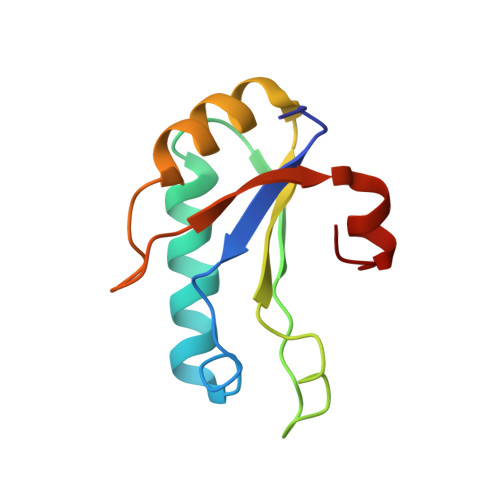
Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP
Selenko, P., Gregorovic, G., Sprangers, R., Stier, G., Rhani, Z., Kramer, A., Sattler, M.(2003) Mol Cell 11: 965-976
- PubMed: 12718882
- DOI: https://doi.org/10.1016/s1097-2765(03)00115-1
- Primary Citation of Related Structures:
1O0P, 1OPI - PubMed Abstract:
The essential splicing factors SF1 and U2AF play an important role in the recognition of the pre-mRNA 3' splice site during early spliceosome assembly. The structure of the C-terminal RRM (RRM3) of human U2AF(65) complexed to an N-terminal peptide of SF1 reveals an extended negatively charged helix A and an additional helix C. Helix C shields the potential RNA binding surface. SF1 binds to the opposite, helical face of RRM3. It inserts a conserved tryptophan into a hydrophobic pocket between helices A and B in a way that strikingly resembles part of the molecular interface in the U2AF heterodimer. This molecular recognition establishes a paradigm for protein binding by a subfamily of noncanonical RRMs.
Organizational Affiliation:
European Molecular Biology Laboratory, Meyerhofstrasse 1, D-69117 Heidelberg, Germany.