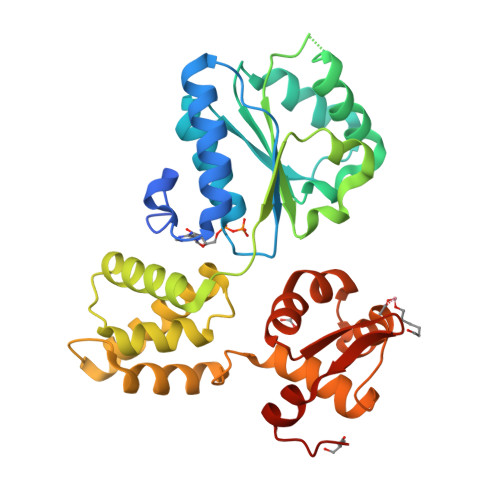
Structure and mechanism of the RuvB Holliday junction branch migration motor.
Putnam, C.D., Clancy, S.B., Tsuruta, H., Gonzalez, S., Wetmur, J.G., Tainer, J.A.(2001) J Mol Biology 311: 297-310
- PubMed: 11478862
- DOI: https://doi.org/10.1006/jmbi.2001.4852
- Primary Citation of Related Structures:
1IN4, 1IN5, 1IN6, 1IN7, 1IN8, 1J7K - PubMed Abstract:
The RuvB hexamer is the chemomechanical motor of the RuvAB complex that migrates Holliday junction branch-points in DNA recombination and the rescue of stalled DNA replication forks. The 1.6 A crystal structure of Thermotoga maritima RuvB together with five mutant structures reveal that RuvB is an ATPase-associated with diverse cellular activities (AAA+-class ATPase) with a winged-helix DNA-binding domain. The RuvB-ADP complex structure and mutagenesis suggest how AAA+-class ATPases couple nucleotide binding and hydrolysis to interdomain conformational changes and asymmetry within the RuvB hexamer implied by the crystallographic packing and small-angle X-ray scattering in solution. ATP-driven domain motion is positioned to move double-stranded DNA through the hexamer and drive conformational changes between subunits by altering the complementary hydrophilic protein- protein interfaces. Structural and biochemical analysis of five motifs in the protein suggest that ATP binding is a strained conformation recognized both by sensors and the Walker motifs and that intersubunit activation occurs by an arginine finger motif reminiscent of the GTPase-activating proteins. Taken together, these results provide insights into how RuvB functions as a motor for branch migration of Holliday junctions.
Organizational Affiliation:
Department of Molecular Biology, Skaggs Institute for Chemical Biology, The Scripps Research Institute, MB 4, 10550 North Torrey Pines Rd, La Jolla, CA 92037, USA.