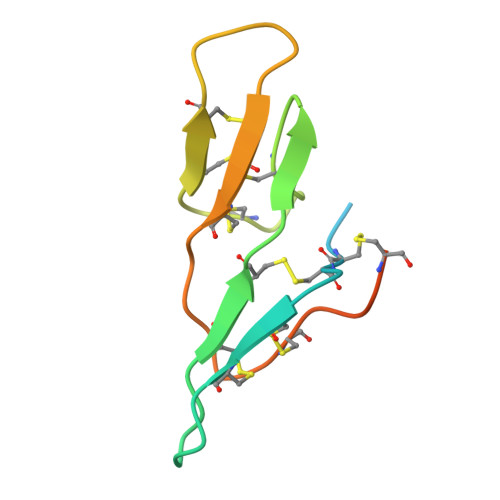
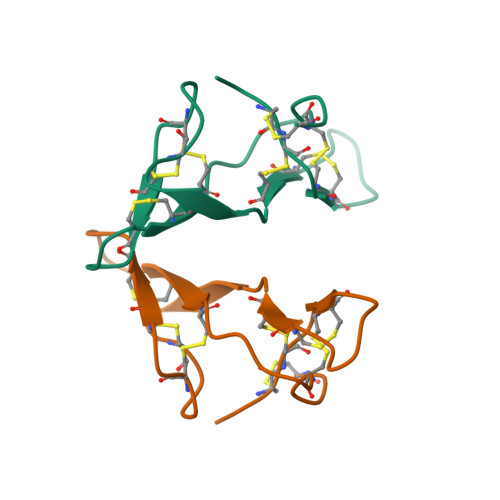
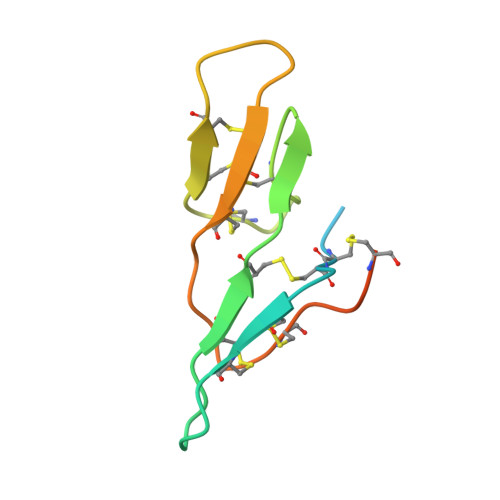
In-House Phase Determination of the Lima Bean Trypsin Inhibitor: A Low-Resolution Sulfur-Sad Case
Debreczeni, J.E., Bunkoczi, G., Girmann, B., Sheldrick, G.M.(2003) Acta Crystallogr D Biol Crystallogr 59: 393
- PubMed: 12554963
- DOI: https://doi.org/10.1107/s0907444902020917
- Primary Citation of Related Structures:
1H34 - PubMed Abstract:
SAD (single-wavelength anomalous diffraction) has enormous potential for phasing proteins using only the anomalous signal of the almost ubiquitous native sulfur, but requires extremely precise data. The previously unknown structure of the lima bean trypsin inhibitor (LBTI) was solved using highly redundant data collected to 3 A using a CCD detector with a rotating-anode generator and three-circle goniometer. The seven 'super-S' atoms (disulfide bridges) were located by dual-space recycling with SHELXD and the high solvent content enabled the density-modification program SHELXE to generate high-quality maps despite the modest resolution. Subsequently, a 2.05 A synchrotron data set was collected and used for further phase extension and structure refinement.
Organizational Affiliation:
Lehrstuhl für Strukturchemie, Georg-August Universität, Göttingen, Germany.