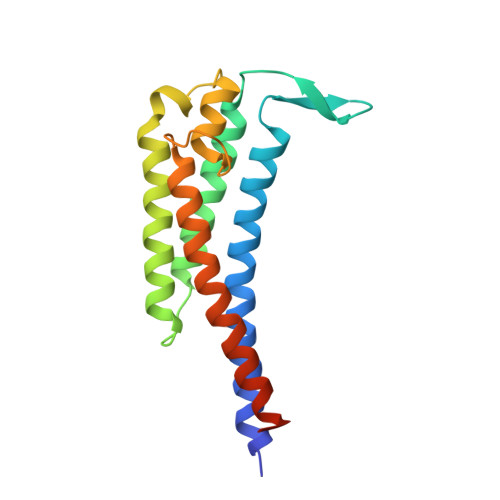
Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.
Kumaran, D., Eswaramoorthy, S., Luft, B.J., Koide, S., Dunn, J.J., Lawson, C.L., Swaminathan, S.(2001) EMBO J 20: 971-978
- PubMed: 11230121
- DOI: https://doi.org/10.1093/emboj/20.5.971
- Primary Citation of Related Structures:
1F1M, 1GGQ - PubMed Abstract:
Outer surface protein C (OspC) is a major antigen on the surface of the Lyme disease spirochete, Borrelia burgdorferi, when it is being transmitted to humans. Crystal structures of OspC have been determined for strains HB19 and B31 to 1.8 and 2.5 A resolution, respectively. The three-dimensional structure is predominantly helical. This is in contrast to the structure of OspA, a major surface protein mainly present when spirochetes are residing in the midgut of unfed ticks, which is mostly beta-sheet. The surface of OspC that would project away from the spirochete's membrane has a region of strong negative electrostatic potential which may be involved in binding to positively charged host ligands. This feature is present only on OspCs from strains known to cause invasive human disease.
Organizational Affiliation:
Biology Department, Brookhaven National Laboratory, Upton, NY 11973, USA.