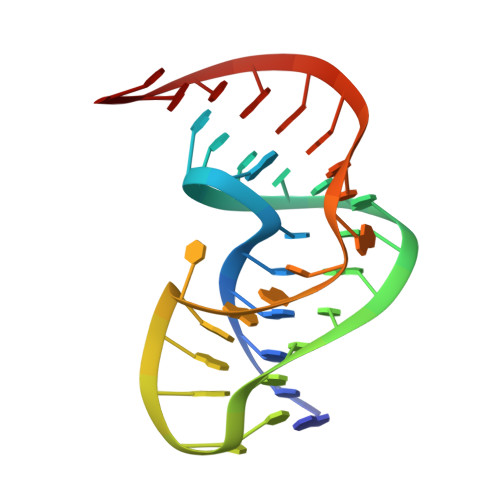
Solution Structure of the Pseudoknot of Srv-1 RNA, Involved in Ribosomal Frameshifting
Michiels, P.J.A., Versleijen, A.A., Verlaan, P.W., Pleij, C.W.A., Hilbers, C.W., Heus, H.A.(2001) J Mol Biol 310: 1109
- PubMed: 11501999
- DOI: https://doi.org/10.1006/jmbi.2001.4823
- Primary Citation of Related Structures:
1E95 - PubMed Abstract:
RNA pseudoknots play important roles in many biological processes. In the simian retrovirus type-1 (SRV-1) a pseudoknot together with a heptanucleotide slippery sequence are responsible for programmed ribosomal frameshifting, a translational recoding mechanism used to control expression of the Gag-Pol polyprotein from overlapping gag and pol open reading frames. Here we present the three-dimensional structure of the SRV-1 pseudoknot determined by NMR. The structure has a classical H-type fold and forms a triple helix by interactions between loop 2 and the minor groove of stem 1 involving base-base and base-sugar interactions and a ribose zipper motif, not identified in pseudoknots so far. Further stabilization is provided by a stack of five adenine bases and a uracil in loop 2, enforcing a cytidine to bulge. The two stems of the pseudoknot stack upon each other, demonstrating that a pseudoknot without an intercalated base at the junction can induce efficient frameshifting. Results of mutagenesis data are explained in context with the present three-dimensional structure. The two base-pairs at the junction of stem 1 and 2 have a helical twist of approximately 49 degrees, allowing proper alignment and close approach of the three different strands at the junction. In addition to the overwound junction the structure is somewhat kinked between stem 1 and 2, assisting the single adenosine in spanning the major groove of stem 2. Geometrical models are presented that reveal the importance of the magnitude of the helical twist at the junction in determining the overall architecture of classical pseudoknots, in particular related to the opening of the minor groove of stem 1 and the orientation of stem 2, which determines the number of loop 1 nucleotides that span its major groove.
Organizational Affiliation:
NSR Center for Molecular Structure, Design and Synthesis, Laboratory of Biophysical Chemistry, University of Nijmegen, The Netherlands.