Structure of the Ser195Ala mutant of human alpha--thrombin complexed with fibrinopeptide A(7--16): evidence for residual catalytic activity.
Krishnan, R., Sadler, J.E., Tulinsky, A.(2000) Acta Crystallogr D Biol Crystallogr 56: 406-410
- PubMed: 10739913
- DOI: https://doi.org/10.1107/s0907444900001487
- Primary Citation of Related Structures:
1DM4 - PubMed Abstract:
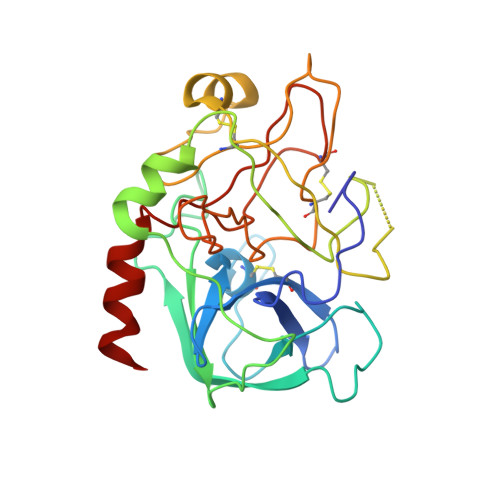
The Ser195Ala mutant of human alpha-thrombin was complexed with fibrinopeptide A(7-22) (FPA) in an effort to describe the (P1'-P6') post-cleavage binding subsites of the fibrinogen-recognition exosite and define more clearly the nature of the Michaelis complex and the scissile peptide bond bound at the catalytic site. The thrombin mutant, however, has residual catalytic activity and proteolysis occurred at the Arg16-Gly17 bond. Thus, the structure of the thrombin complex determined was that of FPA(7-16) bound at the active site, which is very similar to the ternary FPA(7-16)cmk-human thrombin-hirugen complex (r.m.s.d. approximately 0.4 A; Stubbs et al. , 1992). It is further shown by subsidiary experiments that the cleavage is the result of residual catalytic activity of the altered catalytic machinery.
- Department of Chemistry, Michigan State University, East Lansing, MI 48824, USA.
Organizational Affiliation: