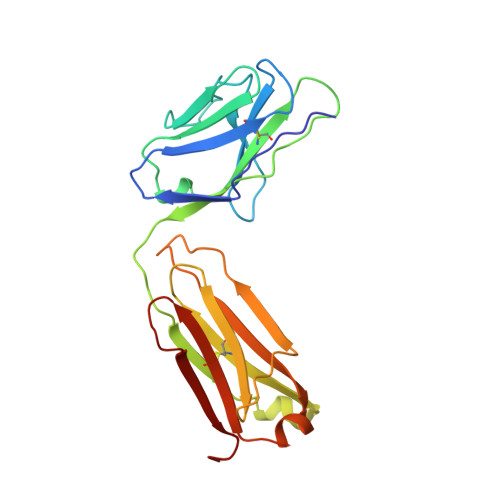
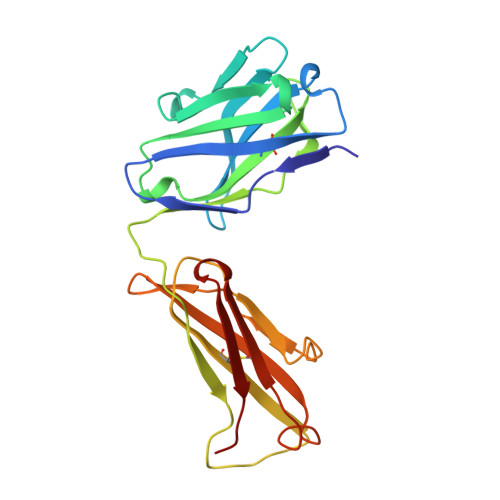
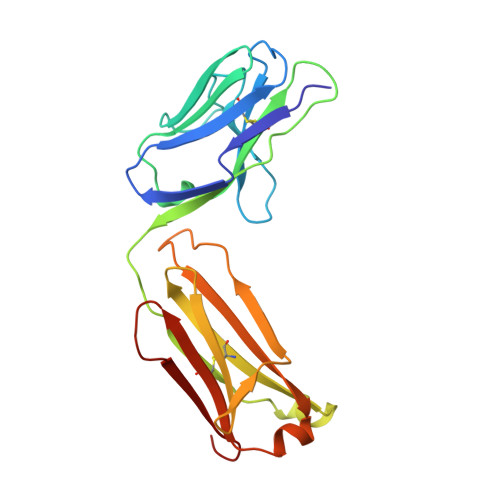
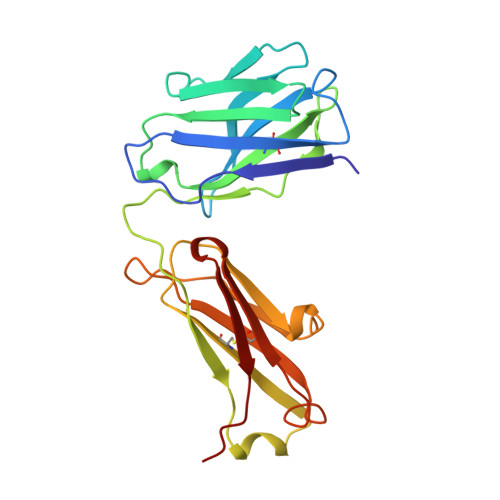
Three-dimensional structure of an idiotope-anti-idiotope complex.
Bentley, G.A., Boulot, G., Riottot, M.M., Poljak, R.J.(1990) Nature 348: 254-257
- PubMed: 1700305
- DOI: https://doi.org/10.1038/348254a0
- Primary Citation of Related Structures:
1CIC - PubMed Abstract:
Serologically detected antigenic determinants unique to an antibody or group of antibodies are called idiotopes. The sum of idiotopes of an antibody constitute its idiotype. Idiotypes have been intensively studied following a hypothesis for the self-regulation of the immune system through a network of idiotype-anti-idiotype interactions. Furthermore, as antigen and anti-idiotypes can competitively bind to idiotype-positive, antigen-specific antibodies, anti-idiotypes may carry an 'internal image' of the external antigen. Here we describe the structure of the complex between the monoclonal anti-lysozyme FabD1.3 and the anti-idiotopic FabE225 at 2.5 A resolution. This complex defines a private idiotope consisting of 13 amino-acid residues, mainly from the complementarity-determining regions of D1.3. Seven of these residues make contacts with the antigen, indicating a significant overlap between idiotope and antigen-combining site. Idiotopic mimicry of the external antigen is not achieved at the molecular level in this example.
Organizational Affiliation:
Unité d'Immunologie Structurale, URA 359 CNRS, Département d'Immunologie, Institut Pasteur, Paris, France.