Architecture and functional regulation of a plant PSII-LHCII megacomplex
Shan, J.Y., Liu, Z.F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Biological assembly 1 assigned by authors.
Macromolecule Content
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II protein D1 | A, UB [auth AA], UC [auth Aa], WA [auth a] | 351 | Spinacia oleracea | Mutation(s): 0 EC: 1.10.3.9 | 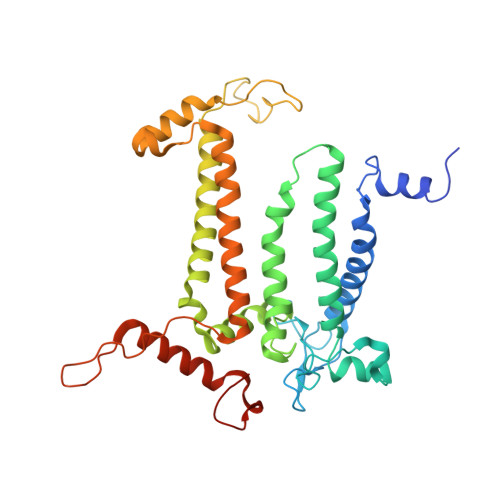 |
UniProt | |||||
Find proteins for P69560 (Spinacia oleracea) Explore P69560 Go to UniProtKB: P69560 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P69560 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Oxygen-evolving enhancer protein 1, chloroplastic | B [auth Oo], JC [auth OO], KB [auth o], LA [auth O] | 332 | Spinacia oleracea | Mutation(s): 0 | 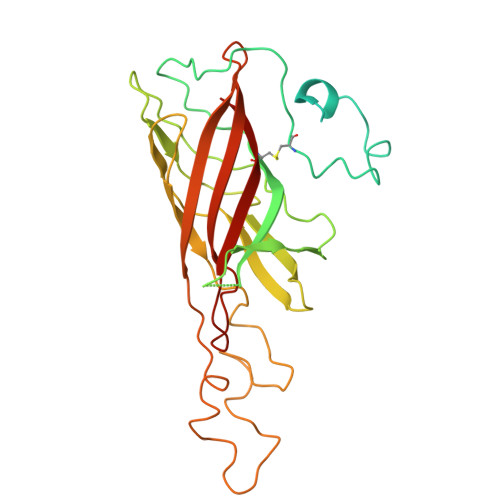 |
UniProt | |||||
Find proteins for P12359 (Spinacia oleracea) Explore P12359 Go to UniProtKB: P12359 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12359 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chlorophyll a-b binding protein, chloroplastic | C [auth Rr], LB [auth r], MC [auth RR], OA [auth R] | 286 | Spinacia oleracea | Mutation(s): 0 | 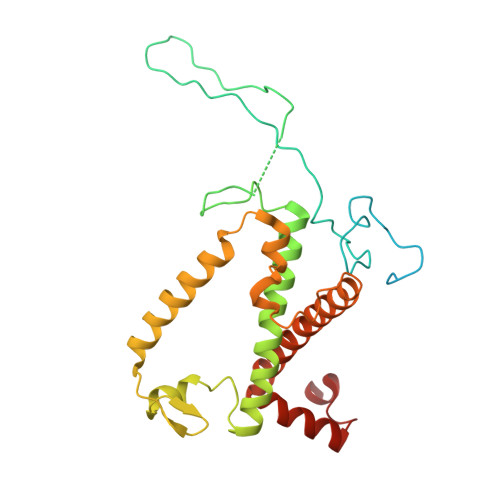 |
UniProt | |||||
Find proteins for A0A9R0HRF2 (Spinacia oleracea) Explore A0A9R0HRF2 Go to UniProtKB: A0A9R0HRF2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A9R0HRF2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chlorophyll a-b binding protein, chloroplastic | D [auth Ss], MB [auth s], NC [auth SS], PA [auth S] | 295 | Spinacia oleracea | Mutation(s): 0 | 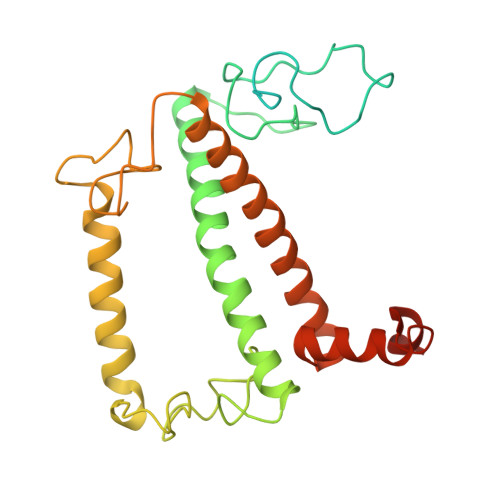 |
UniProt | |||||
Find proteins for A0A9R0IJF3 (Spinacia oleracea) Explore A0A9R0IJF3 Go to UniProtKB: A0A9R0IJF3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A9R0IJF3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein T | E [auth Tt], NB [auth t], OC [auth TT], QA [auth T] | 33 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P61840 (Spinacia oleracea) Explore P61840 Go to UniProtKB: P61840 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61840 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II 5 kDa protein, chloroplastic | F [auth Uu], OB [auth u], PC [auth UU], RA [auth U] | 99 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A9R0J8H2 (Spinacia oleracea) Explore A0A9R0J8H2 Go to UniProtKB: A0A9R0J8H2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A9R0J8H2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center W protein, chloroplastic | G [auth Ww], PB [auth w], QC [auth WW], SA [auth W] | 137 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q41387 (Spinacia oleracea) Explore Q41387 Go to UniProtKB: Q41387 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q41387 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center X protein, chloroplastic | H [auth Xx], QB [auth x], RC [auth XX], TA [auth X] | 117 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A9R0JQ89 (Spinacia oleracea) Explore A0A9R0JQ89 Go to UniProtKB: A0A9R0JQ89 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A9R0JQ89 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chlorophyll a-b binding protein, chloroplastic | 267 | Spinacia oleracea | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for A0A9R0JBE4 (Spinacia oleracea) Explore A0A9R0JBE4 Go to UniProtKB: A0A9R0JBE4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A9R0JBE4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein Z | J [auth Zz], SB [auth z], TC [auth ZZ], VA [auth Z] | 62 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q9M3M6 (Spinacia oleracea) Explore Q9M3M6 Go to UniProtKB: Q9M3M6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9M3M6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chlorophyll a-b binding protein CP24, chloroplastic | K [auth 44], TB [auth 4] | 259 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P36494 (Spinacia oleracea) Explore P36494 Go to UniProtKB: P36494 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P36494 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Oxygen-evolving enhancer protein 2, chloroplastic | KC [auth PP], L [auth Pp], MA [auth P], N [auth p] | 267 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P12302 (Spinacia oleracea) Explore P12302 Go to UniProtKB: P12302 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12302 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Oxygen-evolving enhancer protein 3, chloroplastic | LC [auth QQ], M [auth Qq], NA [auth Q], O [auth q] | 232 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P12301 (Spinacia oleracea) Explore P12301 Go to UniProtKB: P12301 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12301 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chlorophyll a-b binding protein, chloroplastic | P [auth 11], Q [auth 22], S [auth 1], T [auth 2] | 267 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A4F3R4 (Spinacia oleracea) Explore A4F3R4 Go to UniProtKB: A4F3R4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A4F3R4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chlorophyll a-b binding protein, chloroplastic | R [auth 33], U [auth 3] | 264 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for A0A9R0J656 (Spinacia oleracea) Explore A0A9R0J656 Go to UniProtKB: A0A9R0J656 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A9R0J656 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II 10 kDa polypeptide, chloroplastic | V [auth 0], X [auth 00] | 140 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P10690 (Spinacia oleracea) Explore P10690 Go to UniProtKB: P10690 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P10690 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center proteins PsbY, chloroplastic | W [auth 5], Y [auth 55] | 199 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P80470 (Spinacia oleracea) Explore P80470 Go to UniProtKB: P80470 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P80470 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 18 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II CP43 reaction center protein | WB [auth CC], WC [auth Cc], YA [auth c], Z [auth C] | 473 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P06003 (Spinacia oleracea) Explore P06003 Go to UniProtKB: P06003 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06003 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 19 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II D2 protein | AA [auth D], XB [auth DD], XC [auth Dd], ZA [auth d] | 352 | Spinacia oleracea | Mutation(s): 0 EC: 1.10.3.9 |  |
UniProt | |||||
Find proteins for P06005 (Spinacia oleracea) Explore P06005 Go to UniProtKB: P06005 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06005 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 20 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b559 subunit alpha | AB [auth e], BA [auth E], YC [auth Ee], ZB [auth EE] | 83 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P69383 (Spinacia oleracea) Explore P69383 Go to UniProtKB: P69383 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P69383 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 21 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b559 subunit beta | AC [auth FF], BB [auth f], CA [auth F], ZC [auth Ff] | 39 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P60128 (Spinacia oleracea) Explore P60128 Go to UniProtKB: P60128 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60128 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 22 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein H | BD [auth Hh], CC [auth HH], DB [auth h], EA [auth H] | 73 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P05146 (Spinacia oleracea) Explore P05146 Go to UniProtKB: P05146 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P05146 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 23 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein I | CD [auth Ii], DC [auth II], EB [auth i], FA [auth I] | 36 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62103 (Spinacia oleracea) Explore P62103 Go to UniProtKB: P62103 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62103 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 24 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein J | DD [auth Jj], EC [auth JJ], FB [auth j], GA [auth J] | 40 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q9M3L2 (Spinacia oleracea) Explore Q9M3L2 Go to UniProtKB: Q9M3L2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9M3L2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 25 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein K | ED [auth Kk], FC [auth KK], GB [auth k], HA [auth K] | 59 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P12163 (Spinacia oleracea) Explore P12163 Go to UniProtKB: P12163 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12163 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 26 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein L | FD [auth Ll], GC [auth LL], HB [auth l], IA [auth L] | 38 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P60150 (Spinacia oleracea) Explore P60150 Go to UniProtKB: P60150 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60150 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 27 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II reaction center protein M | GD [auth Mm], HC [auth MM], IB [auth m], JA [auth M] | 34 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62112 (Spinacia oleracea) Explore P62112 Go to UniProtKB: P62112 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62112 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 28 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Photosystem II CP47 reaction center protein | VB [auth BB], VC [auth Bb], XA [auth b], YB [auth B] | 508 | Spinacia oleracea | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P04160 (Spinacia oleracea) Explore P04160 Go to UniProtKB: P04160 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04160 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Chinese Academy of Sciences | China | -- |
RCSB PDB Core Operations are funded by the U.S. National Science Foundation (DBI-2321666), the US Department of Energy (DE-SC0019749), and the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM157729.


















