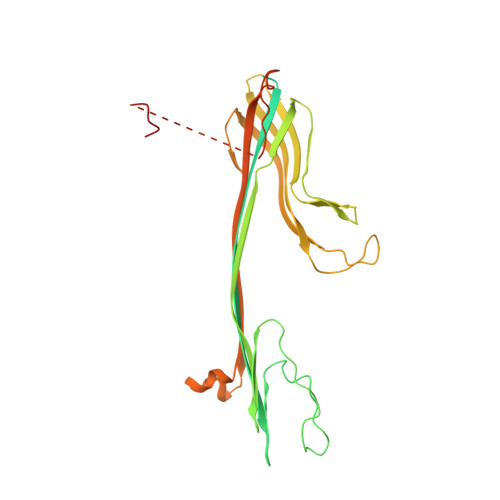
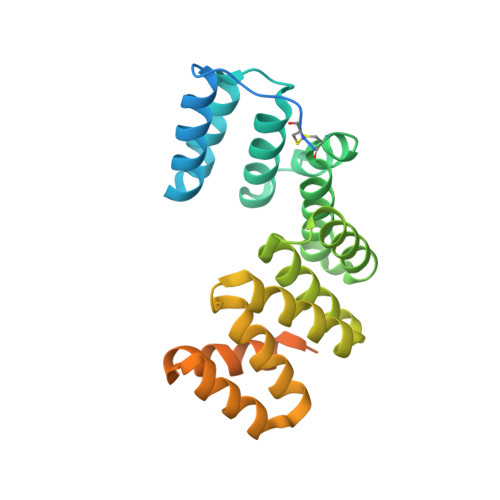
Assembly mechanism of a Tad secretion system secretin-pilotin complex.
Tassinari, M., Rudzite, M., Filloux, A., Low, H.H.(2023) Nat Commun 14: 5643-5643
- PubMed: 37704603
- DOI: https://doi.org/10.1038/s41467-023-41200-1
- Primary Citation of Related Structures:
8ODN - PubMed Abstract:
The bacterial Tight adherence Secretion System (TadSS) assembles surface pili that drive cell adherence, biofilm formation and bacterial predation. The structure and mechanism of the TadSS is mostly unknown. This includes characterisation of the outer membrane secretin through which the pilus is channelled and recruitment of its pilotin. Here we investigate RcpA and TadD lipoprotein from Pseudomonas aeruginosa. Light microscopy reveals RcpA colocalising with TadD in P. aeruginosa and when heterologously expressed in Escherichia coli. We use cryogenic electron microscopy to determine how RcpA and TadD assemble a secretin channel with C13 and C14 symmetries. Despite low sequence homology, we show that TadD shares a similar fold to the type 4 pilus system pilotin PilF. We establish that the C-terminal four residues of RcpA bind TadD - an interaction essential for secretin formation. The binding mechanism between RcpA and TadD appears distinct from known secretin-pilotin pairings in other secretion systems.
Organizational Affiliation:
Department of Infectious Disease, Imperial College, London, SW7 2AZ, UK.