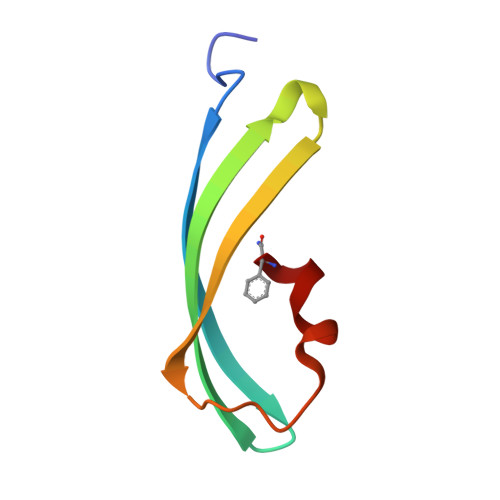
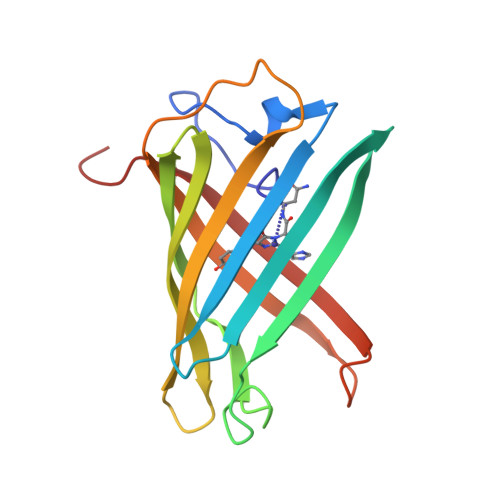
Structural Basis for Photo-Induced Protein Cleavage and Green-to-Red Conversion of Fluorescent Protein Eosfp.
Nienhaus, K., Nienhaus, G.U., Wiedenmann, J., Nar, H.(2005) Proc Natl Acad Sci U S A 102: 9156
- PubMed: 15964985
- DOI: https://doi.org/10.1073/pnas.0501874102
- Primary Citation of Related Structures:
1ZUX, 2BTJ - PubMed Abstract:
Genetically encoded fusion constructs derived from fluorescent proteins (FPs) can be designed to report on a multitude of events and signals in cells, tissues, and entire organs without interfering with the complex machinery of life. EosFP is a novel FP from the scleractinian coral Lobophyllia hemprichii that switches its fluorescence emission from green (516 nm) to red (581 nm) upon irradiation with approximately 400-nm light. This property enables localized tagging of proteins and thus provides a valuable tool for tracking protein movements within live cells. Here, we present the x-ray structures of the green and red forms of WT EosFP. They reveal that formation of the red chromophore is associated with cleavage of the peptide backbone, with surprisingly little change elsewhere in the structure, and provide insights into the mechanism that generates this interesting posttranslational polypeptide modification.
Organizational Affiliation:
Department of Biophysics and General Zoology, University of Ulm, Albert-Einstein-Allee 11, D-89081 Ulm, Germany.