MxB N-Terminus Adopts a Stable alpha-Helix to Engage the HIV-1 Capsid Trimer Interface
Zhu, Y., Rey, S.J., Shen, J., Perilla, R.J., Zhang, P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Currently 9SLY does not have a validation slider image.
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gag polyprotein | 232 | Homo sapiens | Mutation(s): 0 Gene Names: gag | 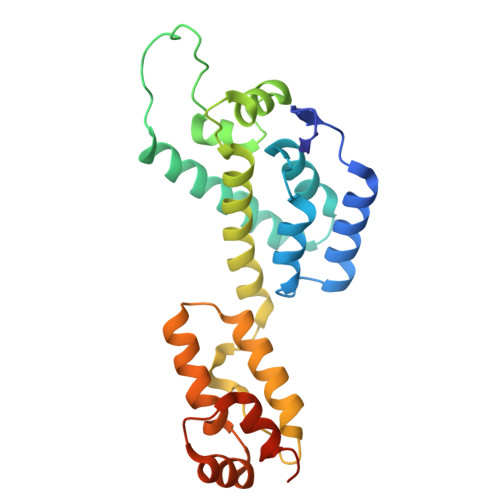 | |
UniProt | |||||
Find proteins for T2CI25 (Human immunodeficiency virus type 1) Explore T2CI25 Go to UniProtKB: T2CI25 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | T2CI25 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
Currently 9SLY does not have a validation slider image.
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | -- |