Ligand efficacy is driven by conformational control of nucleotide exchange at the mu opioid receptor.
Khan, S., Han, G.W., Gati, C.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | A [auth B] | 361 | Homo sapiens | Mutation(s): 0 Gene Names: GNB1 | 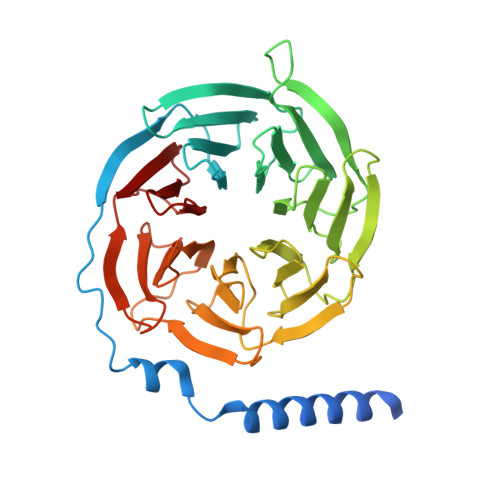 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62873 (Homo sapiens) Explore P62873 Go to UniProtKB: P62873 | |||||
PHAROS: P62873 GTEx: ENSG00000078369 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62873 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | B [auth C] | 71 | Homo sapiens | Mutation(s): 0 Gene Names: GNG2 | 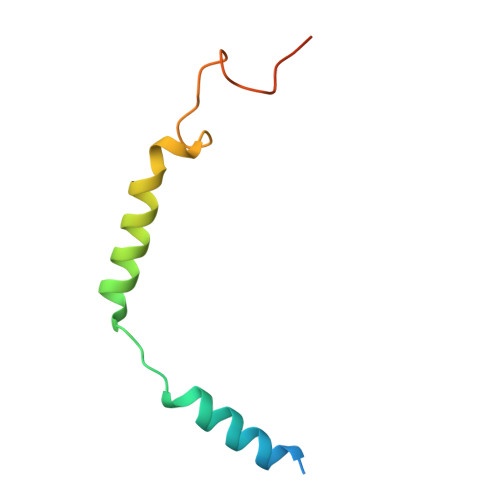 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P59768 (Homo sapiens) Explore P59768 Go to UniProtKB: P59768 | |||||
PHAROS: P59768 GTEx: ENSG00000186469 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P59768 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Soluble cytochrome b562,Mu-type opioid receptor | C [auth R] | 622 | Homo sapiens | Mutation(s): 0 Gene Names: cybC, OPRM1, MOR1 | 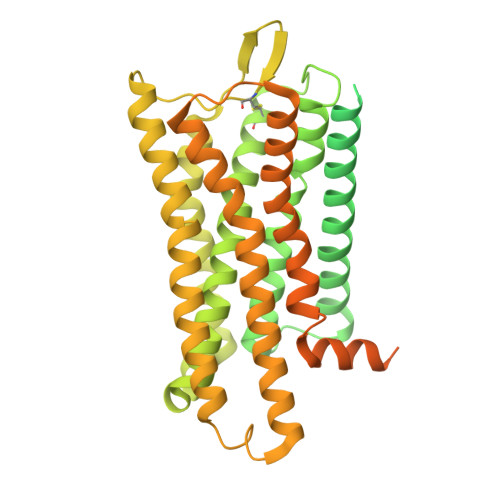 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P0ABE7 (Escherichia coli) Explore P0ABE7 Go to UniProtKB: P0ABE7 | |||||
Find proteins for P35372 (Homo sapiens) Explore P35372 Go to UniProtKB: P35372 | |||||
PHAROS: P35372 GTEx: ENSG00000112038 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | P0ABE7P35372 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(i) subunit alpha-1 | D [auth A] | 354 | Homo sapiens | Mutation(s): 0 Gene Names: GNAI1 EC: 3.6.5 | 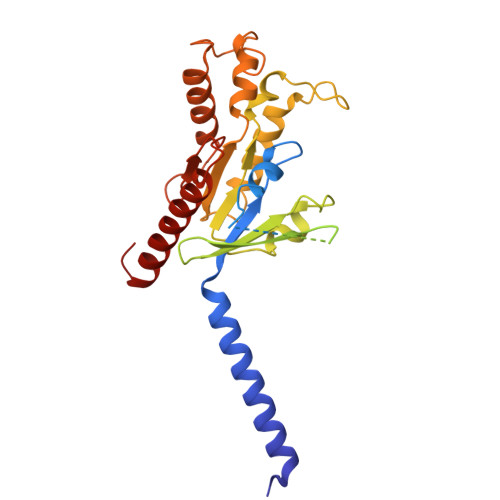 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P63096 (Homo sapiens) Explore P63096 Go to UniProtKB: P63096 | |||||
PHAROS: P63096 GTEx: ENSG00000127955 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63096 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1CMV (Subject of Investigation/LOI) Query on A1CMV | G [auth R] | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-N,N-dimethyl-2,2-diphenylbutanamide C29 H33 Cl N2 O2 RDOIQAHITMMDAJ-UHFFFAOYSA-N |  | ||
| VSN (Subject of Investigation/LOI) Query on VSN | H [auth A] | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine C10 H15 N5 O10 P2 S QJXJXBXFIOTYHB-UUOKFMHZSA-N |  | ||
| CLR Query on CLR | E [auth R], F [auth R] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | v4.3.1 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM144965 |
| National Institutes of Health/National Center for Complementary and Integrative Health (NIH/NCCIH) | United States | AT012075 |