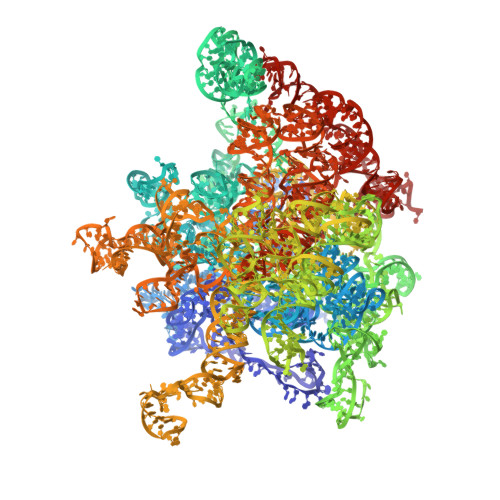
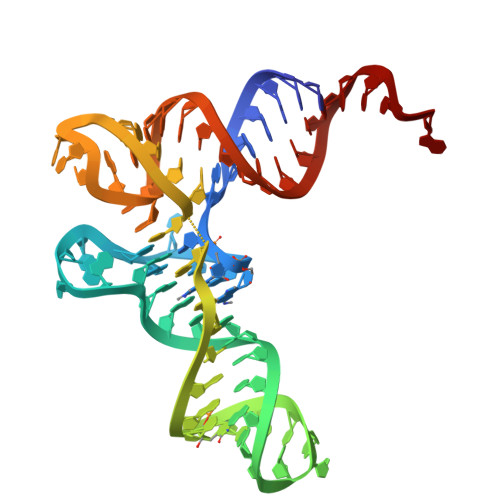
Structural basis of TACO1-mediated efficient mitochondrial translation.
Wang, S., Brischigliaro, M., Zhang, Y., Wu, C., Zheng, W., Barrientos, A., Xiong, Y.(2026) Nat Commun
- PubMed: 41663403
- DOI: https://doi.org/10.1038/s41467-026-69156-y
- Primary Citation of Related Structures:
9OLF, 9PR4, 9PRA, 9PRD, 9PRQ, 9PRX, 9PS0, 9PS7, 9PSI, 9PSM - PubMed Abstract:
Translation elongation is a universally conserved step in protein synthesis, relying on elongation factors that engage the ribosomal L7/L12 stalk to mediate aminoacyl-tRNA delivery, accommodation, and ribosomal translocation. Using in organello cryo-electron microscopy, we reveal how the mitochondrial translation accelerator TACO1 promotes efficient elongation on human mitoribosomes. TACO1 binds the mitoribosomal region typically bound by elongation factor Tu (mtEF-Tu), bridging the large and small subunits via contacts with 16S rRNA, bL12m, A-site tRNA, and uS12m. While active throughout elongation, TACO1 is especially critical when translating polyproline motifs. Its absence prolongs mtEF-Tu residence in A/T states, causes persistent mitoribosomal stalling and premature subunit dissociation. Structural analyses indicate that TACO1 competes with mtEF-Tu for mitoribosome binding, stabilizes A-site tRNA, and enhances peptidyl transfer through a mechanism distinct from EF-P and eIF5A. These findings suggest that bacterial TACO1 orthologs may serve analogous roles, highlighting an evolutionarily conserved strategy for maintaining elongation efficiency during challenging translation events.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT, USA.
Organizational Affiliation: