Structural insights into the engagement of lysophosphatidic acid receptor 1 with different G proteins.
Suzuki, S., Tanaka, K., Kamegawa, A., Nishikawa, K., Suzuki, H., Oshima, A., Fujiyoshi, Y.(2024) J Struct Biol 217: 108164-108164
- PubMed: 39725093
- DOI: https://doi.org/10.1016/j.jsb.2024.108164
- Primary Citation of Related Structures:
9IZF, 9IZG, 9IZH - PubMed Abstract:
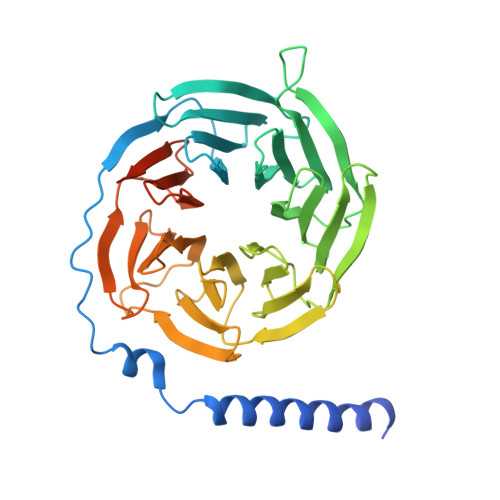
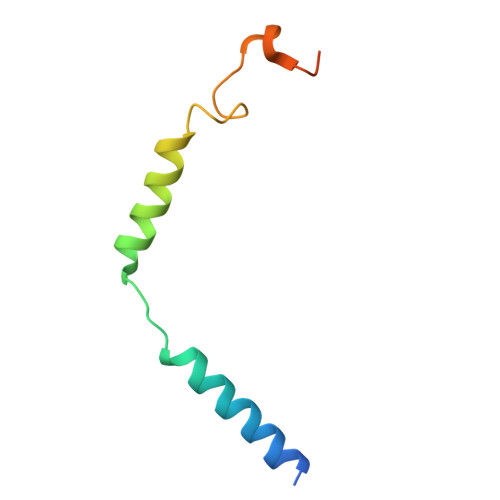
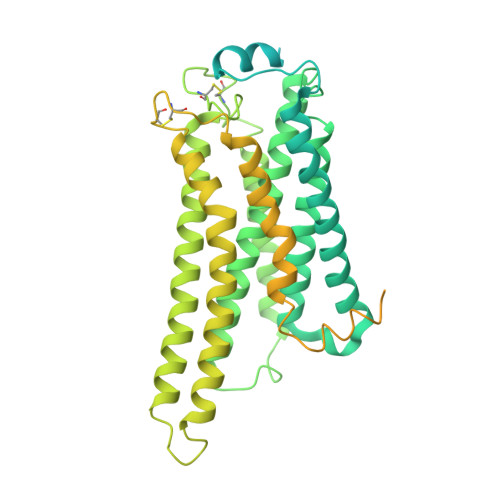
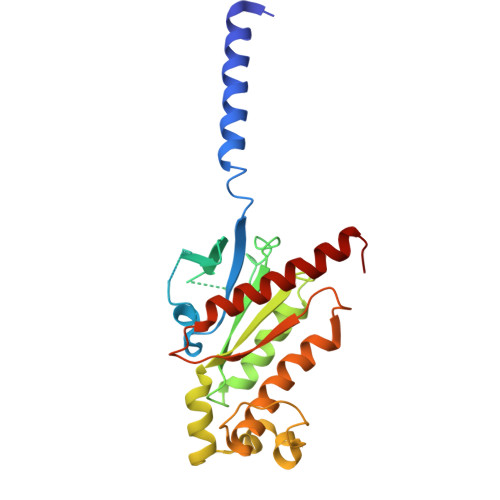
Lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) are bioactive lysophospholipids derived from cell membranes that activate the endothelial differentiation gene family of G protein-coupled receptors. Activation of these receptors triggers multiple downstream signaling cascades through G proteins such as Gi/o, Gq/11, and G12/13. Therefore, LPA and S1P mediate several physiological processes, including cytoskeletal dynamics, neurite retraction, cell migration, cell proliferation, and intracellular ion fluxes. The basis for the G-protein coupling selectivity of EDG receptors, however, remains unknown. Here, we present cryo-electron microscopy structures of LPA-activated LPA1 in complexes with G i , G q , and G 13 heterotrimers . Comparison of the three LPA1-G protein structures shows clearly different conformations of intracellular loop 2 (ICL2) and ICL3 that are likely induced by the different Gα protein interfaces. Interestingly, this G-protein interface interaction is a common feature of LPA and S1P receptors. Our findings provide clues to understanding the promiscuity of G-protein coupling in the endothelial differentiation gene family.
- Advanced Research Initiative, Institute of Integrated Research, Institute of Science Tokyo, 1-5-45 Yushima Bunkyo-ku 113-8510, Tokyo, Japan.
Organizational Affiliation: